Manuscript accepted on : April 25, 2011
Published online on: 28-06-2011
Isolation and Characterization of Bio-Degrading Bacteria from Soil Samples
T. Eswar Ganesh Babu and S. A. Mastan
P.G. Department of Biotechnology, P.G Courses and Research Centre, DNR College, Bhimvaram - 534 202 India.
ABSTRACT: In the present study, a total of four species of biodegrading bacteria namely Bacillus subtilis, Eschericia coli, Staphylococcus aereus and Pseudomonas putida were isolated from soil samples collected from the sites where lead and mercury were discharged. The effect of various physico-chemical parameters on growth of these bacteria was also studied.
KEYWORDS: Biodegradation; Bacteria; Soil Sample
Download this article as:| Copy the following to cite this article: Babu T. E. G, Mastan S. A. Isolation and Characterization of Bio-Degrading Bacteria from Soil Samples. Biosci Biotech Res Asia 2011;8(1) |
| Copy the following to cite this URL: Babu T. E. G, Mastan S. A. Isolation and Characterization of Bio-Degrading Bacteria from Soil Samples. Biosci Biotech Res Asia 2011;8(1). Available from: https://www.biotech-asia.org/?p=9335 |
Introduction
Biodegradation technology uses microorganisms to reduce, eliminate or transform the benign contaminants and their products which are present in soil, sediments, water and air. Evidence of kitchen middens (ancient household garbage dumps) and compost piles dates back to 6000 B.C. demonstrating that some form of biodegradation and bioremediation was practiced by humans since the beginning of recorded history. Biodegradation was used over 100 years ago with the opening of the first biological sewage treatment plant in Sussex, UK, in 1891 however, the word “Biodegradation” is fairly new; first appeared in a peer-reviewed scientific literature in 1987.
Biodegradation can be a cost efficient and reliable method for removing hazardous waste from heavily contaminated sites with organic compounds. For the last decade researchers have also discovered that biodegradation technology can be used at sites contaminated with solvents, explosives, polycyclic aromatic hydrocarbons (PAHs), and polychlorinated biphenyls (PCBs). More recently, researchers have discovered that microbial processes can be used to cleanup radioactive and metallic compounds.
Staphylococcus, Bacillus, Pseudomonas, Citrobacteia, Klebsilla, and Rhodococcus are organisms that are commonly used in biodegradation and bioremediation mechanisms. (Al-Maghrabi et al., 1998). These mechanisms include bioaugmentation, in which microbes and nutrients are added to the contaminated site, and biostimulation, in which nutrients and enzymes are added to supplement the intrinsic microbes of the site. For example Alcaligenes and Pseudomonas have been used in the biodegradation of chromium (Barkay and Colwell, 1983). Likewise, there are records that organisms like Escherichia and Pseudomonas are used in the biodegradation of copper (Barkay and Cowell,1983). Currently, more research is being performed on the use of microbes to degrade metals. It is anticipated that government as well as private industries will invest heavily to clean up polluted environments. This investment validates the need to research the utilization of microbial processes to clean up contaminated sites. In the present paper efforts have been made to isolate and identify the biodegrading bacteria from soil.
Material and Methods
Collection of Soil Samples for Bacterial Strains Isolation
The soil samples containing the bacteria were collected from different areas of Tadaepalli and Mangalagiri, Guntur District of Andhra Pradesh where there was an accumulation of the petroleum spills for Lead deposits and where as for Mercury degradation samples were collected from Seashore of Suryalanka, Baptla, Guntur District, and Andhra Pradesh. The soil sample was collected from a depth of 15 inches depth in to the soil where there was high moisture content, because the Bacteria need moisture content for growth. The soil sample collected was dispensed in the sterile bags and sealed and the sample was brought to the laboratory.
Screening of Lead and Mercury Degrading Bacteria
Soil samples were collected from the sites where lead and Mercury effluents were discharged. Different media like Nutrient Agar, Lauria Bertani agar plates containing Lead and Mercury to a final concentration of 5.0ppm and 7.5ppm respectively. Ten fold serial dilutions of the soil samples were made in distilled water and 0.1ml of 10-5 to 10-9 dilutions were plated on different media. The plates were incubated at 37°C for 24 hours and observed for clearance of degradation surrounding the colonies after incubation. The colonies, which cleared the degradation, were transferred to similar agar plates containing 5.0ppm of Pbcl2 for single colony isolation of Lead degrading organism. In another set of plates 7.5ppm concentration of HgCl2 is used for Single colony isolation. The isolated colonies were streaked on Nutrient agar slants for growth and maintenance. Stock cultures were sub cultured monthly and stored at 4°C. The cultures were confirmed based on morphology by Grams staining and performing various biochemical tests.
Identification of the Bacteria
The isolated bacteria were identified using different staining techniques and various biochemical techniques. Such as Gram Staining, Indole production test, Methyl red and Voges proskauer tests, Gelatin Hydrolysis (Production of Gelatinase), Starch hydrolysis, Oxidase Production and Catalase Activity.
Effect of various factors such as pH , temperature and Nacl concentration were also studied on growth isolated bacteria.
Results
In the present study, a total of four species of bacteria were isolated from soil sample collected from different areas of Tadepalli, Mangalagiri, Suryalanka, Guntur Dt. and Vijayawada Corporation, Andhra Pradesh. The isolated bacterial species were identified as Bacillus subtilis, Escherichia coli, Staphylococcus aureus and Pseudomonas putida. The morphological and biochemical characters exhibited by bacteria are given in the Table 1.
Table 1: Showing Physico-Chemical Parameters of Heavy Metals Degrading Bacteria.
| SS.No | Test | B.subtillis | S.auries | E.coli | P. putida | |
| Microscopic Examination | ||||||
| 11 | Simple staining | Rod Shaped | Cocci | Rod shaped | Rod shaped | |
| 22 | Gram staining | Gram positive Bacilli | Gram positive cocci | Gram Negative Rod | Gram Negative Rod | |
| 33 | Motility Test | Motile | Motile | Motile | Motile | |
| Biochemical Examination | ||||||
| 44 | Indole Production Test | Negative | Positive | Negative | Negative | |
| 55 | Methyl Red test | Negative | Positive | Positive | Negative | |
| 66 | Voges-Proskauer Test | Negative | Negative | Positive | Negative | |
| 77 | Citrate utilization Test | Negative | Negative | Positive | Positive | |
| 88 | Starch Hydrolysis Test | Positive | Negative | Positive | Negative | |
| 99 | Gelatin Hydrolysis Test | Positive | Negative | Negative | Positive | |
| 110 | Catalase Test | Negative | Negative | Positive | Positive | |
| 111 | Caesin Hydrolysis | Positive | Negative | Positive | Positive | |
| 212 | Oxidase Test | Positive | Positive | Positive | Positive | |
Bacillus subtilis
From the microscopic examination it is found that Bacillus subtilis is a rod shaped gram positive bacterium and it is motile. All biochemical characters are identified through various tests which gave positive results to starch hydrolysis test, gelatin hydrolysis test, catalase test, casein hydrolysis and oxidase test and negative results to indole production test, methyl red test, voges proskauer, citrate utilization and catalase negative. (Table 1)
Eshcherichia coli
Morphological examination shows that coli is a gram positive, motile and coccus bacterium. It showed positive results to indole production test, methyl red test and oxidase test and showed negative results to Voges-Proskauer, citrate utilization, starch utilization, gelatin hydrolysis test, catalase test and casein hydrolysis. (Table 1)
Pseudomonas putida
From morphological examination it is clear that Pseudomonas putida is gram negative, motile and rod shaped bacteria. Biochemical examination of putida is given positive results to citrate utilization test, gelatin hydrolysis test, catalase test, casein hydrolysis and oxidase test and negative results to indole production test, methyl red test, voges-proskauer test. (Table 1)
Staphylococcus aureus
Staphylococcus is a gram positive cocci and motile bacteria. Biochemical tests are performed with this strain and it has given positive results to methyl red test, voges Proskauer, citrate utilization, starch hydrolysis, catalase hydrolysis, casein hydrolysis and oxidase test and negative results to indole production test and gelatin hydrolysis test. (Table 1)
Bacillus subtilis culture, 2.E.coli culture, 3.Pseudomonas putida culture, Staphylococcus aureus culture
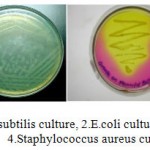 |
Figure 1
|
Effect of various physico-chemical parameters on growth of Bacteria:
Effect of various physico-chemical parameters such as temperature, PH and NaCl were studied on the growth of degrading bacteria involved was carried out during the present investigation. The results obtained indicate the effect of physico-chemical parameters on the growth of the three bacteria involved in degrading lead and mercury, namely, Bacillus substilis, Staphyloccus aureus, and E. coli and P. putida.
Growth pattern of the Bacterial strain
Table 1: Studies on the effect of Physico-chemical parameters (Temperature, pH and NaCl) on the growth of degrading bacteria.
| Factor | Varied Volume | Lead | Mercury | ||
| B.substilis | S.aureus | E.coli | P.putida | ||
| Temperature | 25 | + | + | + | – |
| 28 | + | + | + | – | |
| 30 | + | + | + | + | |
| 35 | + | ++ | ++ | ++ | |
| 37 | + | + | ++ | ++ | |
| 40 | – | + | + | + | |
| 45 | _ | _ | _ | _ | |
| 55 | _ | _ | _ | _ | |
| pH | 2.5 | _ | _ | _ | _ |
| 3.5 | _ | _ | _ | _ | |
| 4.5 | _ | _ | _ | _ | |
| 5.5 | + | + | + | + | |
| 6.5 | ++ | ++ | ++ | ++ | |
| 7 | + | + | + | + | |
| 7.5 | + | + | + | + | |
| 8.5 | + | + | + | + | |
| 9.5 | _ | _ | _ | _ | |
| NaCl (%) | 0 | + | + | + | + |
| 0.5 | + | + | + | + | |
| 2.5 | _ | _ | _ | _ | |
| 5 | _ | _ | _ | _ | |
| 7.5 | _ | _ | _ | _ | |
| 10 | _ | _ | _ | _ | |
Bacillus subtilis
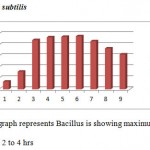 |
Graph 1
|
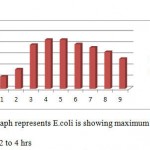 |
Graph 2
|
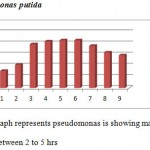 |
Graph 3
|
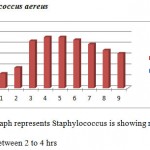 |
Graph 4
|
Above graph represents Staphylococcus is showing maximum growth between 2 to 4 hrs
Effect of temperature in the growth of bacteria
All the bacteria involved in the present study when autoclaved have shown good growth at temperature ranging between 25 to 40 degrees C while they have not shown any growth above 40 degrees C. Good growth rate was observed in between 35 – 37 degrees C in cultures of S. aureus, E.coli, and P. putida.
Effect of pH on growth of bacteria
All four types of bacteria are susceptible to the pH ranging between 5.5 and 8.5 and none of them showed growth below pH 5.5 and above pH8.5. Optimal growth of all the bacteria studied was noted at pH 6.5.
Effect of NaCl on the growth of bacteria
Good growths of S. aureus, B. subtilis, E. coli and P. putida are obtained in 0 to 0.5 percentage of NaCl solution. Growth was not observed in 2.5 and above concentration of NaCl solution.
Discussion
Over the pat one decade, process industries have increased many folds. Industrial uses of metals such as metal plating, tanneries, industrial process utilization metal as catalysts, have generated large amounts of aqueous effluents that contain high levels of heavy metals, these heavy metals include Cadmium, Chromium, Cobalt, Copper, iron, Mercury, Nickel, Silver and Zinc. Metal polluted industrial effluents discleavsed into sewage treatment plants could lead to high metal concentrations in the activated sludge.
Microbial population in metal polluted environment contain microorganisms which have adapted to toxic concentrations of heavy metals and become metal-resistance . At present, metal polluted industrial effluents are mostly treated by the chemical method such as chemical precipitation, electrochemical treatment and ion exchange. These methods provide only partially effective treatment and are costly to implement, especially when the metal concentration is low. Thus biosorption (which is removal using sorption into living/dead organisms) for the removal and recovery of toxic metals from industrial effluents can be economical and effective methods for metal removal. The metal removal ability of microorganisms including bacteria ( Bizily et al.,2000), microalgae and fungi (Bizily et al.,1999)has been studied extensively. The heavy metal removal capacity is higher than those conventional methods. And uptake of heavy metals can be selective (Furukawa and Tonomura., 1972). Microbial cells can also be supplied inexpensively as waste in industrial fermentation processes as well as biological waste water treatment plants ( Bizily et al., 1999).
In the present study, the isolated bacteria exhibited the same biochemical characters described by Buchanan and Gibbon (1987). By virtue of these biochemical characters the isolated bacteria have confirmed up to the species level.
In the present investigation it has been found that different physico – chemical parameters such as temperature, pH and Nacl etc have their influence in the growth of bacteria. Good growth observed at 37°C temperature, while in case of pH 6.5 is optimum for the four species of bacteria. In case of Nacl for the four species of bacteria the concentration is 0 to 0.5 percentage of Nacl was favorable for the good growth of four species of bacteria.
References
- Al-MaghrabiI.M.A,O.Chaalal and M.R.Islam (1998) : Thermophilic bacteria in UAEenvironment can enhance biodegradation and miligate wellbore problems.Proc.SPE. ADIPEC 98 , 228pp. Abu Dhabi.
- Barkay,T and R.R.Cowell (1983) Altered mercury transport in mutant of Psedomonas fluorescens B69.J.Gen.Microbiol,vol.129,p2945-2950.
- Bizily,SP,CL.Rugh and RB.Meagher (2000) Phytodetoxification of hazardous organomercurial by genetically engineered plants.J.Nat.Biotech,vol.18,pp213-217.
- Bizily ,SP.CL.Rugh and A O.Summer and RB.Meagher (1999) Phytoremediation of methylmercury pollution ,merB expression in Araabidopsis thhhhaliana confers resistance of organomercurials,Proc .Nat. Acad.of sci,vol96,pp 6808-6813.
- Buchanan,R.E.and N.E.Gibbon (1974) Bergeys Manual of determinative Bacteriology(8th ) Ed.,Baltimore,Williums and Welkins,New York,pp 2220.
- Chang,JS,Hong,J.Ogunseitan,OA,Olson,BH,(1993) Interaction of mercuric ions with the bacterial growth medium and its effects on enzymatic reduction ofmercury,J.Biotech.Prog.vol.9pp-526-532.
- Furukawa ,K and.K.Tonomura , (1972) Metallic mercury releasing enzyme in mercury resistant Psedomonas,J.Agri.BiolL.Chemi.vol.36.pp217-226

This work is licensed under a Creative Commons Attribution 4.0 International License.





