Manuscript accepted on : October 03, 2011
Published online on: --
Detection of Inducible Clindamycin Resistance in Clinical Isolates of Staphylococcus aureus
Deepali S. Kamble, Swati S. Nale, Dnyaneshwari P. Ghadage, Vrishali A. Muley and Arvind V. Bhore
Microbiology and Smt.Kashibai Navale Medical College and General Hospital. Pune - 411 041 (India).
ABSTRACT: Clindamycin is commonly used in the treatment of Staphylococcus aureus. In vitro routine tests for clindamycin susceptibility may fail to detect inducible clindamycin resistance due to erm genes resulting in treatment failure, thus necessitating the need to detect such resistance by a simple D test on routine basis. 140 Staphylococcus aureus isolates were subjected to routine antibiotic susceptibility testing including oxacillin (1µg) by Kirby Bauer disc diffusion method. Inducible clindamycin resistance was detected by D test, as per CLSI guidelines on Staphylococcus aureus isolates. 76 (54.28%) were Staphylococcus aureus and 64 (45.71%) were Methicillin resistant Staphylococcus aureus. In MRSA 07 (10.93%) isolates showed inducible clindamycin resistance, 08 (12.5%) showed constitutive resistance, 02 (3.1%)showed MS phenotype while in MSSA 02 (2.63%) isolates showed only inducible clindamycin resistance and not the constitutive resistance , 55 (72.36%) showed MS phenotype. Inducible resistance and Constitutive resistance were found to be higher in MRSA as compared to MSSA. The present study showed that, to avoid the therapeutic failure D test must be performed by all laboratories routinely.
KEYWORDS: Clindamycin resistance; Constitutive MLSB phenotype; Inducible MLSB phenotype; MRSA; MS phenotype
Download this article as:| Copy the following to cite this article: Kamble D. S, Nale S. S, Ghadage D. P, Muley V. A, Bhore A. V. Detection of Inducible Clindamycin Resistance in Clinical Isolates of Staphylococcus aureus. Biosci Biotech Res Asia 2011;8(2) |
| Copy the following to cite this URL: Kamble D. S, Nale S. S, Ghadage D. P, Muley V. A, Bhore A. V. Detection of Inducible Clindamycin Resistance in Clinical Isolates of Staphylococcus aureus. Biosci Biotech Res Asia 2011;8(2). Available from: https://www.biotech-asia.org/?p=9673 |
Introduction
Staphylococcus aureus infections are important causes of nosocomial and community acquired infections. Treatment of these infections is a growing problem due to increasing Methicillin resistance among Staphylococci.1 The Macrolide-Lincosamide- Streptogramin B (MLSB) family of antibiotics serve as an alternative treatment option, with clindamycin being the preferred agent due to its excellent pharmacokinetic properties.2 However, widespread use of MLSB antibiotics has led to an increase in number of Staphylococcal strains acquiring resistance to MLSB antibiotics.3 Macrolide resistance may be due to enzymes encoded by a variety of erm genes- MLSB phenotype or active efflux pump encoded by the mrsA gene- MS phenotype. MLSB resistance can be either constitutive (c MLSB) or inducible (i MLSB). 4 In constitutive resistance, in vitro susceptibility tests show resistance to both erythromycin and clindamycin. In inducible resistance, in vitro susceptibility tests show resistance to erythromycin, but susceptibility to clindamycin, unless induced by erythromycin. iMLSB resistance can not be determined by using standard susceptibility test methods, but can be determined by erythromycin- clindamycin disc approximation test (D test).
The aim of the present study was to determine the prevalence of inducible clindamycin resistance in both Methicillin resistant and susceptible strains of Staphylococcus aureus in clinical isolates and also susceptibility pattern of the isolates in our hospital.
Subjects and Methods
The present study was conducted for a period of 2 years from January 2007 to December 2008 and included a total of 140 isolates of Staphylococcus aureus from specimens of Pus/Wound swabs, Respiratory tract infections and Body fluids. The S. arueus were identified by using standard microbiological procedures.5 Antibiotic susceptibility tests were performed by Kirby Bauer disc diffusion method on Mueller Hinton agar plates using Erythromycin (15µg),Clindamycin (2µg),Penicillin(10U),Cefazolin (30µg) Ciprofloxacin(5µg) Oxacillin (1µg), Trimethoprim-Sulfomethaxazole (1.25/23.75µg ),Tetracycline (30µg) Vancomycin ( 30µg) and Teicoplanin (30µg) as per Clinical Laboratory Standards Institute (CLSI) guidelines Methicillin resistance was detected by Oxacillin disc diffusion method.6
To identify i MLSB phenotype, the D test was performed. A lawn culture of the isolate which was adjusted to 0.5 McFarland’s concentration was made on Mueller Hinton agar plate and discs of clindamycin (2µg) and erythromycin (15µg) were placed at a distance of 15mm (edge to edge) as per CLSI recommendations.
Four different phenotypes were interpreted as follows-
D positive (i MLSB phenotype):-isolates showing resistance to erythromycin, while being sensitive to clindamycin with a D shaped zone of inhibition around clindamycin with flattening toward erythromycin disc. (Fig.1) D negative (MSB phenotype):-
No flattening of the clindamycin zone, resistant to erythromycin but susceptible to clindamycin.(Fig.3).
Constitutive resistance(c MLSB phenotype):- Resistant to both erythromycin toclindamycin. (Fig. 2) Sensitive phenotype: – Sensitive to both erythromycin to clindamycin.
 |
Figure 1: D positive (i MLSB phenotype).
|
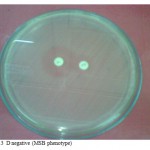 |
Figure 2: Constitutive resistance(c MLSB phenotype).
|
 |
Figure 3: D negative (MSB phenotype).
|
Results
A total of 140 Staphylococci were isolated from various types of clinical specimens. Out of 140 isolates, 76 were Staphylococcus aureus and 64 were Methicillin resistant Staphylococcus aureus (MRSA).
The erythromycin and clindamycin resistance patterns of the isolates based on disc diffusion method are shown in the table- 1.
Table 1:- Distribution of MRSA and MSSA with different resistant phenotypes
| Strains | No. of Isolates (%)
|
iMLSB phenotype
(D +ve) (%)
|
cMLSB phenotype (%)
|
MS phenotype
(D -ve) (%)
|
Sensitive phenotype (%)
|
| MRSA | 64 (45.71)
|
07 (10.93) | 08 (12.5) | 47(73.43) | 02 (3.1) |
| MSSA | 76 (54.28)
|
02 (2.63) | 00 | 10 (13.15) | 64 (84.21) |
| TOTAL | 140
|
09 (6.42) | 08 (5.71) | 57 (40.71) | 66 (47.14) |
cMLSB phenotype (12.5%) was predominant in MRSA. Sensitive phenotype was significantly higher in MSSA (84.21%). No cMLSB phenotype was detected in MSSA.
iMLSB phenotype (6.42%) was slightly higher than c MLSB phenotype (5.71%) among the isolates.
The antibiotic susceptibility patterns of i MLSB isolates are shown in the table.2. All isolates were susceptible to Vancomycin and Teicoplanin while all were resistant to penicillin and Ciprofloxacin. Sensitivity was least to Trimethoprim- Sulfomethaxazole and Tetracycline.
Table 2:- Sensitivity of i MLSB isolates to antimicrobial agents .
| Antimicrobial agents | No. of resistant strains | No.of sensitive strains |
| Oxacillin (1µg) | 07 | 02 |
| Penicillin(10U) | 09 | 00 |
| Cefazolin (30µg) | 07 | 02 |
| Vancomycin ( 30µg) | 00 | 09 |
| Teicoplanin(30µg) | 00 | 09 |
| Ciprofloxacin(5µg) | 09 | 00 |
| Trimethoprim- Sulfomethaxazole
(1.25/23.75µg ), |
07 | 02 |
| Tetracycline (30µg) | 07 | 02 |
Discussion
Due to the increasing Nosocomial MRSA infections which are multidrug resistant treatment option is very limited. Vancomycin due to its high cost and possibility of emergence of resistance; it is not widely used by clinicians.7 Clindamycin due to its tolerability, cost, oral form and good tissue penetration remains alternative treatment option for skin and soft tissue infections. 8 But there are reports of iMLSB resistance in the clinical isolates. 2,3,4,7,8,9,10,11,12 So it is essential to detect the inducible resistance to avoid therapeutic failure in patients.
In our study iMLSB resistance was 6.42%. cMLSB resistance was 5.71%. Similar findings of higher inducible resistance were reported by Angel et al (64% vs. 00%) and Shantala G B et al (24.89% vs. 18.26%).9,10 Deotale et.al (14.5% vs. 3.6%). 11 However in MRSA cMLSB resistance (12.5%) was higher than that of iMLSB resistance (10.93%).Many studies have reported similar results- Gadepalli et al reported 38% cMLSB resistance and 30 % iMLSB resistance. 3 Gupta et al reported 46% cMLSB resistance and 20 % iMLSB resistance. 12 Debdas et al reported 23% cMLSB resistance and 18 % iMLSB resistance.13 Interestingly cMLSB resistance was not seen in MSSA. Similar result was reported by Angel et al only.
The antibiotic susceptibility patterns of i MLSB isolates in this study showed that treatment options are very limited due to multidrug resistant strains. So clindamycin is the preferred drug for the treatment and for that D test must be performed to avoid therapeutic failure.
In conclusion, to avoid the therapeutic failure, the D test must be performed by all laboratories routinely while reporting the sensitivity against clindamycin in staphylococcal infections.
References
- Boucher H.W and Corey G.R. Epidemiology of Methicillin-Resistant Staphylococcus aureus. Clin Infect Dis 2008; 46: S 344–9.
- Fiebelkorn K. R. Crawford S. A. McElmeel M. L., Jorgensen J. H. Practical Disk Diffusion Method for Detection of Inducible Clindamycin Resistance in Staphylococcus aureus and Coagulase-Negative Staphylococci. J Clin Microbiol, Vol. 41; 2003: 4740–4744.
- Gadepalli R, Dhawan B, Mohanty S, Kapil A, Das B.K, Chaudhry. Inducible clindamycin resistance in clinical isolates of Staphylococcus aureus.Indian J Med Res 2006; 123: 571-3.
- Woods C.R. Macrolide-Inducible Resistance to Clindamycin and the D-Test. Pediatr Infect Dis J 2009; 28: 1115–1118.
- Koneman’s color Atlas and Textbook of Diagnostic Microbiology.6thedition. (Lippincott Williams and Wilkins)
- Clinical and Laboratory Standards Institute. Performance Standards for antimicrobial susceptibility testing; 17th informational supplement vol-27 no.1. Clinical and Laboratory Standards Institute; 2007.
- Pai V, Rao V.I, Rao S.V. Prevalence and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus (MRSA) isolates at a tertiary care hospita in Mangalore, South India. J lab physicians.2010; 2(2): 82-84.
- Levin T.P, Suh B, Axelrod P, Truant A.L and Fekete T. Potential clindamycin resistance in clindamycin susceptible, erythromycin resistant Staphylococcus aureus: report of a clinical failure. Antimicrob Agents Chemother . 2005; 49(3) : 1222- 24.
- Angel M.R, Balaji V,Jaj P, Brahmadathan K.N, Mathews M.S. Prevalence of Inducible clindamycin resistance in Gram positive organisms in a Tertiary care centre. Indian J of Med Microbiol.2008; 26(3): 262-64
- Shantala G.B, Shetty A.S,Rao R.K, Vasudeva and Nagarathnamma T. Detection of Inducible clindamycin resistance in clinical isolates of Staphylococcus aureus by disc diffusion induction test. Journal of Clinical and Diagnostic Research.2011;vol-5 (1): 3637.
- Deotale V, Mendiratta D.K, Raut U, Narang P. Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. Indian J of Med Microbiol.2010; 28(2):124-6.
- Gupta V, Datta P, Rani H, Chander J. Inducible clindamycin resistance in Staphylococcus aureus: A study from North India. J Postgrad Med. 2009; 55:176-9
- Debdas D and Joshi S. Incidence of clindamycin resistance in clinical isolates of Staphylococcus aureus.J Infect Dev Ctries 2011 ; 5(4):316-317

This work is licensed under a Creative Commons Attribution 4.0 International License.





