How to Cite | Publication History | PlumX Article Matrix
Characterization and Identification of Seed Storage Protein of Twelve Lettuce Cultivars
Sameer Hasan Qari1, Ehab Abdel-Razik Kamel1 and Hameda El Sayed2
1Biology Department, Aljamom University College, Umm Al-Qura University, P.O. Box: 2064, Makkah, Saudi Arabia.
2Biology Department, Faculty of Applied Science, Umm Al Qura University, Makkah, Saudi Arabia.
Corresponding Author E-mail: sqarinet@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2350
ABSTRACT: We used SDS-PAGE to evaluate and characterize the protein patterns of seed storage proteins in 12 lettuce cultivars. Total protein content of lettuce seeds in all cultivars did not show any significant difference. Results of SDS- PAGE pattern of a few protein bands were up regulated whereas some other bands showed down regulation. The identified protein patterns may be used protein marker for lettuce cultivars. The seed storage protein analyses helps in characterization and identification of diversity in lettuce crop varieties, cultivars and their wild varieties and also provides information on phylogenetic relationship of the accessions. It is also known that variation in protein bands provide information on the relationship among the used seeds. SDS-PAGE of seed protein using Tris-glycine buffer was performed to study the relationships within 12 cultivars of Lactuca sativa L. (lettuce). In the present study, SDS-PAGE was used to investigate and characterize the protein patterns of seed storage proteins in order to find protein bands as markers for cultivar characterization. Datawereanalyzedby clustering method and similarity coefficients using NTSYSpcversion2.02i. ThevalidityofclusteranalysisofSDS-PAGE seed proteins profile datawasdiscussed.
KEYWORDS: SDS-PAGE; Seed Protein; Lettuce; (Lactuca sativa); Cultivars; Numerical Analysis; Cluster Method; Relationships
Download this article as:| Copy the following to cite this article: Qari S. H, Kamel E. A, Sayed H. E. Characterization and Identification of Seed Storage Protein of Twelve Lettuce Cultivars. Biosci Biotech Res Asia 2016;13(4). |
| Copy the following to cite this URL: Qari S. H, Kamel E. A, Sayed H. E. Characterization and Identification of Seed Storage Protein of Twelve Lettuce Cultivars. Biosci Biotech Res Asia 2016;13(4). Available from: https://www.biotech-asia.org/?p=17012 |
Introduction
The genus Lactuca L. is the one of the economically and medicinally important genera of Asteraceae family, subfamily Cichorioideae, tribe the Lactuceae, lant genetic resources comprise all agricultural crops and their wild relatives of valuable traits1. The genus Lactuca L. belonging to the Asteraceae family, is widely distributed in different geographical and ecological areas. Lettuce and most of the other species of the genus Lactuca L. have been cultivated for their economic and medicinal importance. This review summarizes recent knowledge of the application of biochemical (isozymes) and molecular technologies in Lactuca germplasm in order to better understand the genetic variation, interspecific relationships, taxonomy and breeding as a basis for further research studies. This is in order to better understand and improve breeding programmers and biology of crops for future use in agriculture and food security. Hence, advanced molecular genetic technologies including biochemical and molecular markers, have been developed to overcome those limitations of morphological and cytological traits2,3,4,5,6. Seed germination is a complex process that is influenced by many environmental factors, such as light, temperature, and moisture7. Recently, the genome of Lactuca sativa cv wassequenced2. Salinas.Genetic variation studies are vital for providing information for propagation, taxonomy, disease resistance, and breeding programs as well as conservation and utilization of Lactuca genetic resources. Genetic diversity can be evaluated based on morphological, cytogenetic, biochemical and molecular markers8,9,10,11,12. However, the evaluation of genetic variation based on morphological and cytological traits has the disadvantages of being affected by both genetic and environmental factors and may not provide an accurate measure13.
Lettuce (Lactuca sativa) is a temperateannual or biennial plant of the daisy family Asteraceae. It is most often grown as a leaf vegetable. In many countries, it is typically eaten cold, raw, in salads, sandwiches, hamburgers, tacos and in many other dishes. In some places, including China, lettuce is typically eaten cooked and use of the stem is as important as use of the leaf. Both the English name and the Latin name of the genus are ultimately derived from lac, the Latin word for “milk” referring to the plant’s milky juice14. Mild in flavour, it has been described over the centuries as a cooling counterbalance to other ingredients in a salad. The earliest depiction of lettuce is in the carvings at the temple of Senusret at Karnak, where he offers milk to the god Min, to whom the lettuce was sacred. Lettuce was considered an aphrodisiac food in Ancient Egypt and appears as such in the contendings of Horus and Seth. Later, Ancient Greek physicians believed lettuce could act as a sleep-inducing agent. The Romans cultivated it, and it eventually made its way to the Papal Court at Avignon, France15.
There are six commonly recognized cultivar groups of lettuce which are ordered here by head formation and leaf structure; there are hundreds of cultivars of lettuce selected for leaf shape and color, as well as extended field and shelf life, within each of these cultivar groups: (1) Butterhead (L. sativa var.capitataL.) forms loose heads. Its leaves have a buttery texture. Butterhead cultivars are most popular in Europe. Popular varieties include Boston, Bibb, Buttercrunch, and Tom Thumb. (2) Chinese lettuce (L. sativa var.asparaginaBaily.) types generally have long, sword-shaped, non-head-forming leaves, with a bitter and robust flavour unlike Western types, for use in stir-fried dishes and stews. (3) Crisphead, also called Iceberg, forms tight, dense heads that resemble cabbage. They are generally the mildest of the lettuces, valued more for their crunchy texture than for flavour. Cultivars of iceberg lettuce are the most familiar lettuces in the USA. The name Iceberg refers to the crisp, cold, clean characteristics of the leaves. (4) Loose-leaf (L. sativa var.crispaL.) has tender, delicate and mildly flavored leaves. This group includes oak leaf and lollorosso lettuces. (5) Romaine (L. sativa var.romana Lam.), also called Cos, grows in a long head of sturdy leaves with a firm rib down the center. (6) Summer Crisp, also called Batavian, forms moderately dense heads with a crunchy texture. This type is intermediate between iceberg and loose leaf types.
Lettuces contain antioxidants and Vitamin K, romaine and loose leaf lettuce contain five to six times the Vitamin C and five to ten times the Vitamin A of iceberg. Romaine and butterhead lettuce are good sources of folate. Lettuce naturally absorbs and concentrates lithium16.The taxonomicstatusofcultivatedlettuce(LactucasativaL.), theboundaries among L. sativaandcloserelativesandtheboundariesofthegenus LactucaL., itselfhave beenthesubjectofcontroversyamongtaxonomistsfor many decades.
Plant storage proteins can be classified into two classes; seed storage proteins (SSPs) and vegetative storage proteins (VSPs). SSPs are a set of proteins that accumulate at high levels in seeds during the late stages of seed development, whereas VSPs are proteins that accumulate in vegetative tissues such as leaves, stems and tubers, depending on the plant species. SSP genes were classic targets for work on plant molecular biology17.
The abundant expression of plant storage proteins in seeds allowed for easy detection of the gene transcripts and cDNA cloning during research on plant molecular biology in late 70’s to early 80’s. Characterization of germplasm using biochemical fingerprinting has got special attention due to its increased used in crop improvement and the selection of desirable genotypes for breeding crops. The use of genetic markers and protein profiling has also been successfully used to resolve the taxonomic and evolutionary problems of several crop plants18,19,20,21,22.
Seed protein is highly stable, being unaffected by environmental conditions23.Thus electrophoretic banding patterns of total seed protein as revealed by polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS-PAGE) have provided a valid source of taxonomic evidences and were used to address taxonomic relationships at the generic and specific levels, for example Trifolium24, Sesbania25, the genus Vicia26,the genus Lathyrus27,Lens esculenta28, Hordeum vulgare29, andPsidium30,Rosaceae31,Cruciferae32, Raphanus L.33,Campanulaceae33,Gentiana L. and GentianellaMoench,34, canola35,Onobrychis36and Hordeum Vulgare L. 37.
The seed storage protein analyses help in identification and characterization of diversity in crop varieties, cultivars and their wild varieties and provides information on phylogenetic relationship of the accessions. It is also known that variation in protein bands provide information on the relationship among the used seeds collected from various geographical regions38,22,39. There are different amounts of storage proteins in all plant seeds. They play two main roles including nitrogen and energy source and defense against insects and pathogens such as bacteria and fungi.
The aim of this study is to summarize recent knowledge of the seed storage protein analysis can be a useful tool for identification of species, varieties and cultivars, in the present study, SDS-PAGE was used to investigate and characterize the protein patterns of seed storage proteins in 12 cultivars of Lactuca sativa L. (lettuce) to find protein bands as markers for cultivar characterization. Datawereanalyzedby clustering method and similarity coefficients using NTSYSpcversion2.02i. ThevalidityofclusteranalysisofSDS-PAGE seed proteins profile datawasdiscussed.
Materials and Methods
Plant materials
In the present study, sample of 12 cultivars representingLactuca sativa L. collected from different botanic garden and from the ministry of agriculture in Egypt were used Table (1).
Table 1: The Names of Lettuce Plant (Lactuca sativa)Cultivar and The Sources in This Study
| No; | Scientific Name | CV. | Type | Source |
| 1 | Lactuca sativa var. capitataL. | Great Lakes | Cabbage, Butter Head or Headed | 659 (WS) U S A |
| 2 | Lactuca sativa var. capitataL. | All The Year Round | Cabbage, Butter Head or Headed | Italy |
| 3 | Lactuca sativa var. longifoliaLam. | Local | Cos or Romaine | Egypt |
| 4 | Lactuca sativa var. longifoliaLam. | Romaine | Cos or Romaine | Egypt |
| 5 | Lactuca sativa var. crispaL. | Salad Bowl | Leafy or Curled | U S A |
| 6 | Lactuca sativa var. crispaL. | Simpson’s Curled | Leafy or Curled | U S A |
| 7 | Lactuca sativa var. capitataL. | Butter crunch | Cabbage, Butter Head or Headed | U S A |
| 8 | Lactuca sativa var. capitataL. | Lattuga | Cabbage, Butter Head or Headed | Egypt |
| 9 | Lactuca sativa var. capitataL. | Great Lakes | Cabbage, Butter Head or Headed | Egypt |
| 10 | Lactuca sativa var. capitataL. | Great Lakes | Cabbage, Butter Head or Headed | Bonn – Germany |
| 11 | Lactuca sativa var. longifoliaLam. | Romaine | Cos or Romaine | 206 Marseille – France |
| 12 | Lactuca sativa var. longifoliaLam. | Romaine | Laitue Romaine verteMaraichère | France |
Methods of Analysis
Protein Extraction and Electrophoresis
Seed proteins of the studied samples were analyzed using cont-SDS PAGE based on the method of Weber & Osbon40. The electrophoretic analysis was carried out using Tris-glycine (pH 8.2) as an extraction buffer. 0.1 g of seeds was mixed with an equal weight of pure, clean, sterile fine sand and powdered using a mortar and pestle. Extraction was performed overnight. After centrifugation at 15.000 rpm for 10 min, the supernatant was firstly mixed with equal volume of Tris-HCl pH 6.8 (as a digestion buffer) and boiled for 5 min in water path41, then the supernatant was taken for loading on 12% polyacrylamide gels. Electrophoresis was carried out in a Tris-glycine buffer pH 8.3 at 150 V for 2 – 2.5 h, using a low molecular weight protein mixture as a marker in each run. Gels were then stained in Comassie brilliant blue R-250 for 2 h, distained, photographed and the number of bands revealed in each gel lane were counted and compared with each other. Gel Pro-Analyzer software version 2.0 was used to determine the molecular mass (MM) of each protein band.
Numerical Analysis
The data of SDS-PAGE seed protein banding patterns werescoredaspresent (1) and absence (0). Thedatawasanalyzed tocomparevarioussimilaritycoefficientsandclusteringmethodsandtostudy therelationshipsamongthecultivarsdetail. Clustering methodsand similarity coefficients were testedusingtheproceduresSIMQUAL, SAHNandTREEfromtheprogramNTSYSpcversion2.02i42.
Results and Discussion
SDS-PAGE Electropherogram of the Storage Seed Proteins
Seed protein analysis was carried out on 12 cultivars of lettuce represented the four type of lettuce and the electropherograms produced from seed protein analysis using Tris-glycine buffer revealed great polymorphism among these cultivars as illustrated in Table (2) and Fig. (1). A total number of 73 protein bands were observed within the studied cultivars (57 unique bands and 16 polymorphic bands). Each cultivar was characterized by the presence of unique bands (ranged from 3 bands, recorded in cv. no; 2&3 to 7 bands observed in cv. no; 1). Cultivar no; 10 was found to have six bands and all of them were unique for this cultivar. Cv. no; (11) was found to have the highest number of bands (10), while the lowest number of bands (6) was observed in the sample of cv. no; (10).
Table (2): The molecular weights of protein bands extracted in Tris-glycine Buffer of the Studied different lettuce Plants for 12 cultivars.
| Polymorphism | OTU`s | Mol. Weights | No; | |||||||||||
| 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | 1 | |||
| Unique | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 71.579 | 1 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 62.255 | 2 |
| Polymorphic | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 56.846 | 3 |
| Unique | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 56.662 | 4 |
| Polymorphic | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 56.479 | 0 |
| Unique | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 55.931 | 6 |
| Polymorphic | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 55.750 | 7 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 55.210 | 8 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 54.321 | 9 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 53.795 | 10 |
| Unique | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 49.925 | 11 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 46.184 | 12 |
| Polymorphic | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 45.736 | 13 |
| Polymorphic | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 45.440 | 14 |
| Polymorphic | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 45.293 | 15 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 44.357 | 16 |
| Unique | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 41.276 | 17 |
| Polymorphic | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 40.249 | 18 |
| Polymorphic | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 40.105 | 19 |
| Polymorphic | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 39.961 | 20 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 39.817 | 21 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 36.918 | 22 |
| Polymorphic | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 36.522 | 23 |
| Unique | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 36.260 | 24 |
| Unique | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 36.000 | 25 |
| Unique | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 35.545 | 26 |
| Polymorphic | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 35.245 | 27 |
| Polymorphic | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 34.947 | 28 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 34.799 | 29 |
| Unique | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 34.652 | 30 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 32.655 | 31 |
| Unique | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 32.380 | 32 |
| Unique | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 32.106 | 33 |
| Unique | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 31.835 | 34 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 31.566 | 35 |
| Polymorphic | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 31.433 | 36 |
| Unique | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 31.300 | 37 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 31.167 | 38 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 26.785 | 39 |
| Unique | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 26.660 | 40 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 25.984 | 41 |
| Unique | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24.111 | 42 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 23.554 | 43 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 22.796 | 44 |
| Unique | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 22.322 | 45 |
| Polymorphic | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 22.063 | 46 |
| Polymorphic | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 21.960 | 47 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 21.908 | 48 |
| Unique | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 21.857 | 49 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 20.141 | 50 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 16.883 | 51 |
| Unique | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16.833 | 52 |
| Unique | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 16.783 | 53 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 16.733 | 54 |
| Unique | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16.634 | 55 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 16.486 | 56 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 16.437 | 57 |
| Polymorphic | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 16.389 | 58 |
| Unique | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16.340 | 59 |
| Unique | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16.051 | 60 |
| Unique | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15.581 | 61 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 15.535 | 62 |
| Unique | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14.902 | 63 |
| Unique | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13.958 | 64 |
| Unique | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13.876 | 65 |
| Unique | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13.793 | 66 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 13.753 | 67 |
| Unique | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 13.590 | 68 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 13.390 | 69 |
| Unique | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13.310 | 70 |
| Unique | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13.271 | 71 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 12.730 | 72 |
| Unique | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 09.570 | 73 |
| 9 | 10 | 6 | 9 | 8 | 9 | 8 | 9 | 8 | 7 | 8 | 9 | Total no; of bands | ||
| 4 | 6 | 6 | 6 | 4 | 4 | 5 | 5 | 4 | 3 | 3 | 7 | Number of unique bands | ||
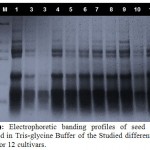 |
Figure 1: Electrophoretic banding profiles of seed proteins extracted in Tris-glycineBuffer of the Studied different lettuce Plants for 12 cultivars. |
The highest molecular weight protein band (71.579 KDa) among the studied samples was recorded in cv. no; (12), while the lowest one (09.570 KDa) was detected in the cv. no;(5).
Seed protein banding patterns as revealed by SDS-PAGE produces reproducible band pattern (profile) when proteins are prepared in a standard method and hence have valid value in taxonomic purposes. Consequently, proteins with identical electrophoretic mobility are deemed to represent the same unit character. Therefore characters derived from seed proteins have been utilized in plant taxonomy at different levels to construct phonetic classifications43,44,45. Hence they can be considered as traits to study genetic variation among the plant taxa. However, for the study of the taxonomic relationships among species and higher taxonomic ranks more valid assessment should necessarily be obtained when these data are compiled with other lines of evidence from morphology and cytology.
Numerical Analysis
The phenogram produced by the analysis of the studied cultivars based on coding of 73 attributes obtained from Tris-glycine extracted proteins are shown in Fig. (2). This phenogram shows that the examined samples have a total similarity coefficient of about 1.52. At this level, cv. no. (1) was split off from the other cultivars, then at 1.48 level cv. no. (11) was split off from the remaining cultivars. At the levels 1.43 and 1.40, cv. no. (9 & 10) were also split off from the remaining cultivars (respectively).
At the level 1.38, the two cultivars (5 & 6) were separated together in a small group from the other cultivars, then they distinguished from each other at the 1.35 level. At the level 1.322, the two cultivars (4 & 12) were separated together from the other cultivars and then distinguished from each other at the 1.31. At the level 1.313, cv. no; (3) was split off from the remaining cultivars. Then at the level 1.29, cv. no; (8) was separated from the remaining cultivars. Finally, at the level of 1.22, cultivars number (2 & 7) were found to be close to each other.
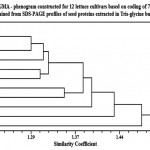 |
Figure 2 |
Electrophoresis of proteins is a powerful tool for identification of genetic diversity and the SDS-PAGE is particularly considered as a reliable technology because seed storage proteins are highly independent of environmental fluctuations46,47. Seed protein patterns can also be used as a promising tool for distinguishing cultivars of particular crop species48,49. However, only a few studies indicated that cultivar identification was not possible with the SDS-PAGE method50. The SDSPAGE is considered to be a practical and reliable method for species identification51.
According to the results of the SDS-PAGE, the overall pattern of seed storage-proteins showed the diversity of lettuce cultivars. The electropherogram produced from seed protein analysis of the 12 cultivars revealed great polymorphism. The diversity in seed storage proteins has also been reported by Khan et al.52for wheat varieties. Moreover, identification of three wheat genotypes including ILC-195, CM-2000 and CM-98/99 has also been reported by protein markers53.
Since in mature seeds, type and amount of proteins are more constant than other plant tissues54therefore, the SDS-PAGE pattern of seed storage proteins of pistachio showed polymorphism based on difference in protein intensity among genotypes. The presence or absence of protein bands has also been applied for detection of polymorphism of Brassica cultivars55.
The present investigation revealed variation in different cultivars of lettuce seeds with regard to their total seed protein profiles. These results of SDS-PAGE seed protein patterns show of a few protein bands (6-10) and identified protein patterns may be used protein marker for lettuce cultivars. Regarding inter specific variation among cultivars this investigation revealed some variations. The genetic affinities within cultivars of the same species generally corroborated the morphological analysis. Similar to our finding the result of differentiation of yellow sarson and brown seeded types of Brassica clearly separated the yellow seeded and brown seeded varieties by SDS-PAGE56. However, we can conclude that, SDS-PAGE can reveal the differences among seed storage proteins of lettuce cultivars.
References
- Bisht,V. K. Purohit, V. Medicinal and Aromatic Plants Diversity of Asteraceae in Uttarakhand. Nature and Science, 2010; 121 -128.
- Lavelle, D.; Wang, Z.; Song, C.; Kozik, A.; Froenicke, L.; Truco, M.; Beitel, C.; Xu, X.; Yang, B.;Michelmore, R.: Sequencing the genomes of cultivated Lactuca sativa and its wild progenitor, L. serriola. In: The Largest Ag-Genomics Meeting in the World. California: USA; 2013.
- Jourdan, N.; Martino, C.; El-Esawi, M.; Witczak, J.;Bouchet, P.E.; d’ Harlingue, A.; Ahmad, M. Blue-light dependent ROS formation by arabidopsis cryptochrome-2 may contribute towards its signaling role. Plant Signal Behav., 2015; 10 (8): e1042647. DOI:10.1080/15592324.2015.104264.
- Consentino, L.; Lambert, S.; Martino, C.; Jourdan, N.; Bouchet, P. E.; Witczak, J.; Castello, P.; El-Esawi, M.; Corbineau, F.; d’Harlingue, A.; Ahmad, M. Blue-light dependent reactive oxygen species formation by Arabidopsis cryptochrome may define a novel evolutionarily conserved signaling mechanism. New Phytol. 2015; 206:1450-1462.
CrossRef - El-Esawi, M.; Glascoe, A.; Engle, D.; Ritz, T.; Link, J.; Ahmad, M. Cellular metabolites modulate in vivo signaling of Arabidopsis cryptochrome-1. Plant Signal Behav. 2015; 10 (9). DOI: 10.1080/15592324.2015.1063758
CrossRef - El-Esawi MA. Taxonomic relationships and biochemical genetic characterization of Brassica resources: Towards a recent platform for germplasm improvement and utilization. Annu Res Rev Biol.,2015; DOI:10.9734/ARRB/2015/20645
CrossRef - Bewley, J.D.; Bradford, K.J.; Hilhorst, H.W.M.; Nonogaki, H. (3rd ed): Seeds: Physiology of Development, Germination and Dormancy, 3rd ed. New York: Springer,2013; 312-320.
CrossRef - El-Esawi MA, Sammour R. Karyological and phylogenetic studies in the genus Lactuca L. (Asteraceae). Cytologia. 2014; 79:269–75.
CrossRef - El-Esawi MA. Assessing the genetic diversity, phylogenetic relationships, and disease resistance genes in Irish Brassica oleracea species. PhD Thesis, Dublin Institute of Technology, Ireland,2012.
- El-Esawi, M.; Bourke, P.; Germaine, K.; Malone, R.Assessment of morphological variation in Irish Brassica oleracea Species. JAS. 2012a; 4(10):20-34.
- El-Esawi, M.; Germaine, K.; Malone, R. Assessing the genetic diversity and relationships in Irish Brassica oleracea species based on microsatellites markers. Proceedings of the Fifth Saudi Science Conference, Umm Al-Qura University, Saudi Arabia (SSC5′, April 16-18, 2012b).
- Sammour R.; Badr S.; Mustafa A.; El-Esawi M. Genetic variation within and among some Lactuca spp. based on karyotype analysis. Applied Cell Biology (ACB),2013; 2(4):136-43.
- Basu, A.; Ghosh, M.; Meyer, R.; Powell, W.; Basak, S.; Sen, S. Analysis of genetic diversity in cultivated jute determined by means of SSR markers and AFLP profiling. Crop Sci. 2004; 44:678-685.
CrossRef - Simpson, D.P: (5th ed). Cassell’s Latin Dictionary (5th ed.). London: Cassell Ltd,1979.
- Grigson, J.: The Vegetable Book. London: Penguin,1978.
- Hullin, R.P.; Kapel, M.; Drinkall, J.A. The lithium contents of some consumable items. International Journal of Food Science & Technology,2007; 4(3): 235-240.
CrossRef - Ehsanpour, A.A.; Shojaie, B.; Rostami, F. Characterization of seed storage protein patterns of four Iranian Pistachios using SDS-PAGE. Natural Science,2010; 2(7): 737-740.
CrossRef - Ladizinsky, G.; Hymowitz, T. Seed protein electrophoresis in taxonomic and evolutionary studies. TAG Theoretical and Applied Genetics,1997; 54(4): 145-151.
CrossRef - Khan, M.K. Production and utility of chickpea (Cicer arietinum L.) in Pakistan. Progressive Farming,1990; 10(6): 28-33.
- Murphy, R.W.; Sites, J.W.; Buth, D.G.;Haufler, C.H: Protein isozymes electrophoresis. In: Molecular Systematic. Hillis, D.H. and Moritz, C., Eds., Sinnauer Association, Massachusetts: Sunderland,1990; 45-126.
- Nakajima, K: Biotechnology for crop improvement in Japan. In: Biotechnology Application in Agriculture in Asia and Pacific, Published by Asian Productivity Organization:1994; 87-107.
- Ghafoor, A.; Gulbaaz, F.N.; Afzal, M.; Ashraf, M. and Arshad, M. Inter-relationship between SDS-PAGE markers and agronomic traits in chickpea (Cicer arietinum L.). Pakistan Journal of Botany,2003; 35(4): 613-624.
- Harborne, J.B.; Turner, B.L (3ed): Plant chemo-systematices, New York:Academic press, 1984; 581-593.
- Badr, A. Electrophoretic studies of seed proteins in relation to chromosomal criteria and the relationships of some taxa of TrifoliumL. Taxon,1995; 44: 183-191.
CrossRef - Badr, A.; Abou-El-Enain, M.M.; El-Shazly, H.H. Variations in seed protein electrophoretic pattern and species relationships in Sesbania. Proceedings of Sixth Egyptian Botanical Conference, Cairo University, Giza, November 24-26. 1998; 3: 493-501.
- Kamel, E.A.R.; Al-Mashad, A.A.A. Electrophoretic studies of seed proteins and the relationships of some species of the genus Vicia L. FABIS Newsletter.,1999; 42: 5-11.
- Badr, A.; El-Shazly H.H.; Abou El-Enain, M. Seed protein diversity and its implications on the relationships in the genus Lathyrus L. (Fabaceae). Proc. 1st Conf. Biol. (ICBS) Fac. Sci., Tanta Univ., 7-8 May,2000; 1: 333-346.
- Hassan, H. Z. Genetic fingerprinting and relationships of some lentil (Lens esculentaMoench.) cultivars based on protein and DNA polymorphism. The 8th International Conference of Union of Arab Biologists, Fayuom Univ., Fac. Sci., 4-7 Nov.,2001; 11-31.
- El-Rabey, H. A., A. Badr, R. Schaffer-Pregl, W. Martin and F. Salamini,. Speciation and species separation in Hordeum L. (Poaceae) resolved by discontinuous molecular markers. Plant Biol. 2002; 4: 567–575.
CrossRef - Hassan, H.Z.; El-Shrif, A.H.; Wafaa T.S. Molecular genetic fingerprinting of some guava (Psidiumguajava L.) cultivars based on SDS-protein, isozymes and RAPD analyses. Bull. Fac. Assiut Univ.,2002; 31 (1-D): 259-282.
- Dowidar, A.E.; Kamel, E.A.; Ahmed, A.H.M.; Loutfy, M.H.A. and Hafez, H.H. L. Studies on The Rosaceae II-SDS-PAGESeed Protein Electrophoresis and Its Significance in The Taxonomy of The Family. Pakistan J. Biol. Sci.,2003; 6(21): 1820-1829.
CrossRef - Kamel, E.A.; Hassan, H.Z. and Ahmed, S.M. Electrophoretic characterization and the relationship between some Egyptian Cruciferae. J. Biol. Sci.,2003; 3(9): 834-842.
CrossRef - Kamel, E.A. Seed Protein Diversity and the Relationships within the Campanulaceae. J. Bot.,2005; 34(2): 507-526.
- Kamel, E. A.; Elwan, Z. A. and Loutfy, M. H. A. On the taxonomy of Gentiana L. and GentianellaMoench. (Gentianaceae): evidence from seed coat morphology and seed protein electrophoresis. J. Bot.,2006;186-198
- Afiah, S.A.; Abdelsalam, A.Z.E.; Kamel, E.A.; Dowidar, A.E. and Ahmed, S.M. Molecular genetic studies on canola crosses under Maryout conditions. 8th African Crop Science Society Conference, October 27th – 31st, 2007.
- Emre, I.; Turgut-balik, D.; Sahin, A. and Kursat, M. Total Electrophoresis Band Patterns of Some Onobrychis Species Growing in Turkey. American-Euraasian J. Agri. & Environ. Sci.,2007; 2(2): 123-126.
- El-Rabey, H.; Abdellatif, K.F. and Khidr, Y.A. Cytogenetical and Biochemical Characterization of Some Egyptian Barley (Hordeum Vulgare L.) Cultivars. Australian Journal of Basic and Applied Sciences,2009; 3(2): 644-651.
- Satija, D.R.; Adrash, B.; Gupta, S.K. and Bala, A. Genetic diversity in relation to protein and protein fractions in chickpea (Cicer arietinum L.). Crop-Improvement, 2002;29(2): 122-135.
- Asghar, R.; Siddique, T.; Afzal, M. Inter and intra-specific variation in SDS-PAGE electrophoregrams of total seed protein in chickpea (Cicer arietinum L.) germplasm. Pakistan Journal of Biological Sciences, 2003;6(24), 1991-1995.
CrossRef - Weber, K.; Osborn, M. The reliability of molecular weight determinations by dodecyl sulphate-polyacrylamide gel electrophoresis. J. Biol. Chem.,1969;244: 4406-4412.
- Gifford, D.J.; Greenwood, J.S.;Bewley, J.D. Deposition of matrix and crystalloid storage proteins during protein body development in the endosperm of Ricinuscommunis L. cv. Hale seed. Plant Physiol. (Lancaster),1982; 69: 1471-1478.
CrossRef - Rohlf, F.J: NTSYSpc numerical taxonomy and multivariate analysis system user guide. Exeter Software, New York:1998; USA.
- Boulter, D: Proteins of legumes. In: Advances in Legume Systematics, R.M. Polhill and P.H. Raven (eds.) Academci press, London, New York:1981; 501-512.
- Smith, B.J. SDS polyacrylamide gel electrophoresis of proteins. In: Walker, J. M. (ed.), Methods in Molecular Biology, Proteins,Vol. 1, New Jersey :Humana Press, 1984; 211-223
CrossRef - Echeverrigaray, S.; Oliveira, A.C.; Carvalho, M.T. and Derbyshire, E. Evaluation of the relationship between lentil accessions using comparative electrophoresis of seed proteins. J. Gen. & Breed., 1998;52: 89- 94.
- Javid, A.; Ghafoor, A.; Anwar, R. Seed storage protein electrophoresis in groundnut for evaluating genetic diversity. Pakistan Journal of Botany, 2004;36(1): 25-29.
- Iqbal, S.H.; Ghafoor, A.; Ayub, N.Relationship between SDS-PAGE markers and Ascochyta blight in chickpea, Pakistan Journal of Botany,2005; 37(1): 87-96.
- Jha, S.S.;Ohri, D. Phylogenetic relationships of Cajanuscajan(L.) Millsp. (pigeonpea) and its wild relatives based on seed protein profiles. Genetic Resources and Crop Evolution,1996; 43(3): 275-281.
CrossRef - Seferoglua, S.; Seferoglua, H.G.; Tekintasa, F.E. and Baltab, F. Biochemical composition influenced by different locations in Uzun pistachio cv. (PistaciaveraL.) grown in Turkey. Journal of Food Composition and Analysis, 2006;19(5): 461-465.
CrossRef - DeVries, I.M.Characterization and identification of Lactucasativa cultivars and wild relatives with SDS-electrophoresis(Lactucasect.Lactuca,Compositae).Genetic Resources and Crop Evolution,1996; 43:193-202.
CrossRef - Gepts, P: Genetic diversity of seed storage proteins in plants. In: Brown, A.H.D., Clegg, M.T.; Kahler, A.L. and Weir, B.S., Eds., Plant Population Genetics, Breeding and Genetic Resources, Sinauer Associates Inc., Sunderland, Massachusetts,1989; 64-82.
- Khan, M.F.; Schumann, E. and Weber, W.E. Characterization of Pakistani wheat varieties for general cultivation in the mountainous regions of Azad Kashmir. Asian Journal of Plant Sciences,2002; 1(6): 699-702.
CrossRef - Zeb, A.; Zahir, A.; Ahmad, T. and Abdumanon, A. Physiochemical characteristics of wheat varieties growing in the same and different ecological regions of Pakistan. Pakistan Journal of Biological Sciences,2006; 9(9): 1823-1828.
CrossRef - Magni, C.; Scarafoni, A.; Herndl, A.; Sessa, F.; Prinsi, B.; Espen, L.;Duranti, M. Combined 2D electrophoretic approaches for the study of white lupin mature seed storage proteome. Phytochemistry,2007; 68(7): 997-1007.
CrossRef - Sadia, M.; Salman, A.M.; Rabbani, M.A.; Pearce, S.R. Electrophoretic characterization and the relationship between some Brassica species. Electronic Journal of Biology,2009; 5(1): 1-4.
- Das, S.; Mukherjee, K.K. Comparative study on seed proteins of Ipomoea. Seed Science and Technology,1995; 23(2): 501-509.

This work is licensed under a Creative Commons Attribution 4.0 International License.





