How to Cite | Publication History | PlumX Article Matrix
Nima Ghahremani Nejad
Department of biology, Faculty of sciences, Islamic Azad University, Tonekabon branch,Tonekabon,Iran.
DOI : http://dx.doi.org/10.13005/bbra/2386
ABSTRACT: Phage therapy for E.coli O157:H7 in animals is considered as a popular research topic. E.coli O157: H7 is a pathogenic potential. It can cause hemolytic uremic syndrome (HUS). Considering this matter, thepresent research aims at analyzing the isolation of E.coli O157:H7 bacteriophages of livestock waste of Mazandaran province using a phage therapy approach. The separation and culture processes of E.coli O157:H7 from livestock wastewaterhave been analyzed in this research. Also, differential tests were used to identify E.coli O157:H7. First, the Lytic bacteriophages of E.coli O157:H7 were isolated from livestock wastewater. Then, both identification and susceptibility tests for bacteria E.coli O157:H7 were done and phages were isolated using various methods. Finally, phage therapy to animal model was conducted. In this study, I have used two methods including disk diffusion and overlay tests. This test can be used for three strains standards including Enteropathogenic E.coli (EPEC), Enterotoxigenic E. Coli (ETEC), Enteroinvasive E. coli (EIEC), and wild E.coli O157:H7. According to the results, 45 samples had lytic phages vs. four different E. coli strains and only 19 samples had significant effect on E.coli O157: H7.Also, it can be concluded that isolated phages affect bacteria in a significant manner (p<0.05).In the current research, the isolated lytic phages removed the infection of E.coli O157:H7 of five groups of mice. As a result, the lytic bacteriophages have the ability to replace antibiotics.
KEYWORDS: E.coli O157:H7; livestock; Mazandaran; mouse; phage therapy
Download this article as:| Copy the following to cite this article: Nejad N. G. Isolation of E.coli O157:H7 Bacteriophages of Livestock Waste of Mazandaran Province (Case Study: A Phage Therapy Approach). Biosci Biotech Res Asia 2016;13(4). |
| Copy the following to cite this URL: Nejad N. G. Isolation of E.coli O157:H7 Bacteriophages of Livestock Waste of Mazandaran Province (Case Study: A Phage Therapy Approach). Biosci Biotech Res Asia 2016;13(4). Available from: https://www.biotech-asia.org/?p=17171 |
Introduction
The World Health Organization (WHO) has declared antimicrobial resistance as one of the greatest threats to human health. There is urgent need for developingthe antimicrobial agents; however, few are in the pharmaceutical pipeline, and the number of new antibacterial drugs was approved for marketing in the United States continue to drop (World Health Organization, 2001). Also, the Infectious Disease Society of America (IDSA) confirmed that the world is in the midst of an emerging crisis of antibiotic resistance for microbial pathogens(Boucher et al., 2009).Hence, E.coli O157:H7 has become a worldwide threat to public health and is one of today’s most troubling food-borne pathogens. Human illnesses with E.coli O157:H7 range from self-limited watery diarrhea and hemorrhagic colitis to life-threatening manifestations such as the hemolytic uremic syndrome and the thrombotic thrombocytopenicpurpura. Cattle and sheep, important domestic ruminants, are the primary reservoirs for this human pathogen and are the most common sources for foodborne, waterborne, and direct-animal-contact infections. Successful strategies to control or reduce the carriage and prevalence of E.coli O157:H7 in live ruminants would reduce the risk of human exposure to this pathogen(Sheng et al., 2006).Antibiotics have been a panacea in animal husbandry as well as in human therapy for decades. The huge amount of antibiotics was used to induce the growth and protect the health. Itled to the evolution of bacteria that are resistant to the drug’s effects. Today, many researchers are working with bacteriophages (phages) as an alternative to antibiotics in the control of pathogens for human therapy as well as prevention, control, and therapy in animal agriculture. Phage therapy and control have fulfilled their promise or potential yet, largely due to several key obstacles to their performance.Current strategies were employed to combat the rise of multi drug resistance organisms, focus on a more judicious use of antibiotics, and methods to restore the indigenous Microbiota (Knoll et al., 2014). However, E.coli O157:H7 has become a worldwide public health concern since it was first identified as a human pathogen in 1982. This pathogen has a very low infectious dose (approximately 10 cells) in humans, and symptoms of infection range from watery diarrhea to hemorrhagic colitis and hemolytic uremic syndrome, and in some cases, death(Rivas et al., 2010).Since ancient times, there have been documented reports of river waters having the ability to cure infectious diseases such as leprosy (Keen, 2012). However in 1896, the British bacteriologist Ernest-Hankin reported antibacterial activity against Vibrio cholera which he observed in the Ganges and Jumna rivers in India. He suggested that an unidentified substance was responsible for this phenomenon and for limiting the outbreakof cholera epidemics. Two years later, Gamaleya, the Russian bacteriologist, observed a similar phenomenon while working with Bacillus subtilis(Adhya et al., 2006).Phages are naturally occurring predators of bacteria. They can be effective antibacterial agents due to their specific role against particular bacterial species and their lack of impact on the other Microflora(Jassim et al., 2014).Bacteriophages are very specific to their hosts, so this minimizes the chance of secondary infections, but antibiotics do target both pathogens and normal flora of patients, which can cause the secondary infections or sometimes superinfections. Also, bacteriophages replicate at the site of infection where they are mostly needed to lyse the pathogens, but antibiotics travel throughout the body and do not concentrate on the site of infection. No side effects have been reported during or after phage application, but resistant bacteria, allergies (sometimes even fatal anaphylactic reaction), and secondary infections are the common side effects of antibiotics treatment .Lastly, bacteriophages are environmentally friendly and are based on natural selection, isolating and identifying bacteria in a very rapid process compared to new antibiotic development, which may take several years, may cost millions of dollars for clinical trials, and may also not be very cost effective(Weber-Dąbrowska et al., 2001).Effective elimination of pathogenic bacteria from gastrointestinal diseases using phage preparation has been demonstrated in multiple experiments that arefocused on the therapeutic use of phages(Marinelli et al., 2012).The therapeutic effect of the phages can be limited to a decrease in the pathogen’s population down to a point at which the immune system can effectively control its reproduction. Several current strategies to combat livestock-associated pathogens such as toxicogenicE.coli, Campylobacter, and Salmonella are direct extensions of “classical” phage therapy approaches in that they focus on targeting the bacteria in animals before slaughter(Sheng et al., 2006).The prevalence of E.coli O157:H7 in cattle has been the subject of much research, with most studies concluding that at any given time the majority of cattle are negative for E.coli O157:H7(La Ragione et al., 2009).So, some studies have cited sheep products as important sources of E.coli O157:H7.Phages specific for E.coli O157:H7 have previously been isolatedfrom human fecal materials or animal manures from bovine,ovine, swine, and chicken, lake or pond water, and sewage(Tomat et al., 2014; Shahrbabak et al., 2013).The clinical syndromes, pathogenic characteristics, the pathobiology of these organisms and the toxins they produce are reviewed in Kruger and Lucchesi (Krüger et al., 2015).The aim of this study was to isolation of E.coli O157:H7 bacteriophages of Livestock waste of Mazandaran province; a phage therapy approach.
Material and Methods
For preparing wildphages and E.coli O157:H7,70 and 30 samples wererespectivelycollected from many Stockyardsin Tonekabon that located in north of Iran. For this, wastewater contains all kinds of microbes like E.coli O157:H7and phages harvested by sterile dishes. Then, three standard strainsEnteroinvasive E.coli(EIECATCC 43893), Enteropathogenic E.coli (EPEC ATCC 43887), and EnterotoxigenicE.coli (ETEC ATCC 35401)provided from Pasteur Institute of Iranwere used to evaluate the phages.
Isolation of bacteria from livestock wastewater
For isolation of E.coli O157:H7, 1 mlof wastewater was solved in 9ml of physiological serum, then a serial dilution (10-7 to 10-1) was prepared, then concentrations of 10-5 and 10-4 were selected for culturing.It should be noted that for ensuring the accuracy of specialantiserum,we tested it by three standards strain and wild E.coli O157:H7, the results indicated agglutination with E.coli O157:H7. It means that our antiserum act in an appropriate manner (Wagner, 1992).
Isolation of specialbacteriophages from livestock wastewater
For isolation of special phages, 15 ml wastewater was prepared and was centrifuged for 25 min in 3500×g, and then supernatant was filtered via 0.45µm. Then 8.5 ml of filtered liquid was harvested to mix by 10 ml of Nutrient broth media. Then 100µl from E.coli O157:H7 was added to it, and was incubated at 37oc in 150 rpm for 18 hours, in shaking station to airing. Then, it was centrifuged again in 10000×g for 15 min, and then it was filtered through 0.45µm. At last supernatant was gathered for assessing phage activity by two methods: diffusion disks and overlay methods(Ferens et al., 2011).
Differential tests for confirmation of E.coli O157:H7
These tests included: growth on Eosin Methylene Blue, Triple Sugar Iron agar, Simmons Citrate,Sorbitol MacConkey agar,SIM media, gram staining, oxidase, catalase, urease,H2S and CO2 production, glucose and lactose fermentation , IMVIC, , MRVP,and special antiserum for strain E.coli O157:H7(World Health Organization, 2001).
Agglutination test by special antiserum for E.coli O157:H7
In this test, the colonies which were isolated from Sorbitol MacConkey agar media were suspected to E.coli O157:H7 (colorless colonies/sorbitol-negative). They wereselected for agglutination test by E.coli O157:H7 special antiserum. In this method,atfirst, we pilled one droplet normal saline (as control) in right side of slide and one droplet antiserum in left side. Then, we added some E.coli O157:H7 colonies to each droplet.Then we mixed them completely. Then we shook slide for 60 second slowly for observing agglutination process. Formation of agglutination means that existence of E.coli O157:H7.
Detecting E.coli O157:H7 and E.coli O157:H7Special Bacteriophages
Finally, the strain of E.coli O157:H7 wasdetected by using of Sorbitol MacConkey agar and special antiserum. As well, the effective bacteriophages vs. E.coli O157:H7was detected by disk diffusion and Overlay tests. All isolated phage sampleswere tested on three E.colistandard strains (ETEC, EPEC,and EIEC) and wild E.coli O157:H7.
Effect of different antibiotics on E.coli O157:H7
Disks diffusion test contain Tetracycline, Ampicillin and Gentamycin are used to evaluate the sensitivity of wild E.coli O157:H7.For this purpose, wild E.coli O157:H7 bacteria were cultured in Mueller-Hinton media and relevant antibiotics placed on the media and for 48 hourswere incubated at 37oC and then the plates were examined.
Disk diffusion testcontainspecific bacteriophage
In this method, fresh disks were put into final supernatants for 24 hours, and then were incubated at 37oC. To determine the effect of isolated phages, the disks were dried and were put on Nutrient agarmedia(that newly cultured byE.coli O157:H7) by 2cm distance, then wereincubated for 24 hours at 37oC(Payne et al., 2003).
Overlay
Overlay culture was used for this test. First, we cultured E.coli O157:H7 onNutrient agar. Then 400 µl final filtered liquid was piled on that cultured media. Then, 3 ml of Nutrient agar as semi-solid (0.75%) wasadded to it, then plates were incubated for 18-24 hours, at 37oC (Ibid).
Animal stage
For evaluation the effect of phages on bacteria, we divided all mice to six groups, a control group as well as five treated groups. Treated groups include: 1-5. So, groups 1-4 each groups were collected from a different region in north of Iran.Also, fifth group was a mixed of groups 1-4.The isolated lytic phages were those four samples with maximum effect vs. E.coli O157:H7 out of initial samples, so these sampleshad been detected by their diameter in disk diffusion method. These samples were tested on male6-weeks oldmice for 14 days on six groups of by three repeats.The treating of animals lasted 2 weeks. In the first day, all mice (control and others)received 100 µl of wildE.coli O157:H7 liquid (1×107cfu/ml), then after 2 hours, all mice except control group received100 µl phages by two methods: intraperitoneal injection and gavaging,2-times by 3 hours interval (1×108pfu/ml). For preventing digestion of phages in mice gastric acidity, we used calcium carbonate (0.25%) during gavaging. From first day to fourteen, the mice received phages by the same manner.
At day 14, 1 gr stool of micewascollected for testing the existence of E.coli O157:H7. The dilutions of stools (10-3 and 10-4) were prepared and were cultured on EMB, then they were tested by: gram staining, oxidase, urease, IMVIC, MRVP, growth on the Simmons Citrate,Triple Sugar Iron agar, SIM, Sorbitol MacConkey agar, media and special antiserum to evaluate the existence of E.coli O157:H7.Also, the overlay method tested on stools of mice, for evaluating existence of phages(Chapman et al., 2000; Thorpe et al., 2002).
Results
Culture mediums and Biochemical tests
The isolated bacteria from wastewater was cultured on Eosin Methylene Blue media, indicated green metal shining in Eosin Methylene Blue that is a feature of E.coli O157:H7.
In catalase test, from biochemical tests, the samples produced the bulb of air that is an indication of positive-catalase feature.Also, produced blue from oxidase test showed that the samples contained the negative-oxidase bacteria.Indeed, in microscopic observation, the shape of the bacterial sample was negative-gram bacillus.In Triple Sugar Iron agar media, the isolated bacteria changed colorof media to yellow (acid-acid) due to fermenting of glucose and lactose as well as gas formation, but no H2S produced(Figure 1).In SIM media, formation of red ring on surface was observed, that is an indicator of positive-Indole feature of bacteria (using Kovacs reagent). The movement of bacteria observed (it means it have been a flagellated bacteria).Also, no black color was observed that means that no H2S produced, it means the bacteria are H2S-Negative(Figure 1).In IMVIC test, regard to MR test (by methyl red indicator), constant red color on the surface is an indicator of acid formation due to lowering pH under 4.4, so the test is positive.In VP test (by Voges-Proskauer indicator) red color during 15 minute after adding the indicators is an indicator of positive feature, because the existence of di-acetyl is an indicator of acetoinbut the isolated bacteria did not red color, that means the bacteria is positive-MR.In citrate test (Simons citrate media), the bacteria did not change the media color and no growth was observed, it means that they were not able to use citrate as carbon source (negative-citrate); so we know that formation of blue color after 24-48 hours is an indication of citrate consumption and positive featuring. So biochemical results validated that the samples contained E.coli; following table is showed the results of biochemical test on sample (Table1).
 |
Figure 1: Triple Sugar Iron agar media (A), SIM media (B) |
Table 1: The results of biochemical tests and growth on SIM and TSI media
| TSI | SIM | |||||
| H2S | Gas formation | Acid-acid | Indole-formation | H2S Formation | Movement | |
| – | + | + | + | – | + | |
| Other biochemical | IMVIC | |||||
| Oxidase | catalase | Urea hydrolysis | VP | MR | Indole | Citrate |
| – | + | – | – | + | + | – |
Special antibody test
Colonies isolated from Sorbitol MacConkey agar media was agglutinated, that agglutination is an indication of existence of E.coli O157:H7 (Figure 2).
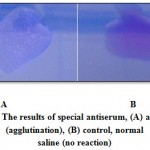 |
Figure 2: The results of special antiserum, (A) antiserum (agglutination), (B) control, normal saline (no reaction) |
Growth on Sorbitol MacConkey agar media
The isolated bacteria produced colorless colonies on Sorbitol Macconkey agar that is a feature of E.coli O157:H7. Because E.coli O157:H7 is not able to fermentation of sorbitol in Sorbitol MacConkey agar, so it produces the colorless colonies, therefore, it can be distinguish from other strains of E.coli that produce pink colonies.
Phages stage
For isolation of E.coliO157:H7 special phages, the number of 70 samples were gathered from livestock wastewater in Tonekabon city.In this study we used two methods disk diffusion and overlay tests. In this test were usedthreestrains standard:EPEC, ETEC and EIEC and wild E.coli O157:H7. Forty five from 70 samples had lytic phages vs. four different E. coli strains, only 19 samples had significant effect on E.coliO157: H7 (Table2).
Table 2: The results of phages isolation from all samples
| Type of phages | Number of Samples |
| Lytic phages effective on all four strains | 11 |
| Lytic phages effective on EPEC | 5 |
| Lytic phages effective on EPEC and ETEC | 10 |
| Lytic and special phages effective on E. coli O157:H7 | 19 |
Disk diffusion test
This test was done for all 70 samples and against all three standard strains as well as for wild E.coli O157:H7. The results from disk diffusion test revealed that the diameter of halosformed by phages was between10-11 mm (Figure 3).Nineteen samples had lytic and special effect against E.coli O157:H7, but no effect on other strains, so these phages are special for E.coli O157:H7.
 |
Figure 3: Evaluation of phage’s effect on E. coli O157:H7 using disk diffusion method |
Overlay method
The results in this method indicated some plaques were formed dueto interaction of bacteriophagesand E.coli O157:H7 on Nutrient agar media.These plaques formed due toinhibitory effect of bacteriophages on E.coli O157:H7.It means that the samples hadenough special bacteriophages against E.coli O157:H7(Figure 4).
 |
Figure 4: The effect of Phage on E. coli O157:H7 using overlay method and plaque formation |
Effect of different antibiotics on E.coli O157:H7
Disks diffusion test contain Tetracycline, Ampicillin and Gentamycin areused to evaluate the sensitivity of wild E.coli O157:H7. This test indicated that isolated E.coli O157:H7 had different sensitivity to different antibiotics. This test indicated that wildE.coli O157:H7 was resistant to Tetracycline but sensitive to isolated special phages and some antibiotics such as Ampicillin and Gentamycin, in thisstudy (Table 3).
Table 3: Evaluation of antibiotics on isolated E. coli O157:H7
| Antibiotics | Effects | Diameter |
| Tetracycline | No sensitive | – |
| Ampicillin | Sensitive | 14mm |
| Gentamycin | Sensitive | 15mm |
The results of phage therapy
Five phage samples have included most effective among all of them against E.coli O157:H7 (based upon halos diameter in diffusion disk method and number of plaque in overlay) were selected for phage therapy stage. This stage lasted for 14 days, so at fourteenth day the excretion of animal gathered for assessing whether have any E.coli O157:H7, as well as inthe same day, we tested existence of bacteriophages from excretion of mice (by disk diffusion and overlay methods). In this stage, in the control group (only received bacteria) showed low-movement as well as diarrhea after 5th days, then inflammation in tail and face and severe diarrhea after 8th days, so at days 10, 12 and 13th, we observed death of mice. In five different groups of experiment, for example first and fourth groups, one of the animal’s death at day 12th due to diarrhea, vomiting and inflammation-Bleeding in tail and face area. In the other groups (2,3and especially mixed phage group), no death is observed (due to special antibacterial effect of isolated phages). So according these results, we can say that the phages could survive infected mice by E.coli O157:H7 from given death, significantly (p<0.05%).
Approval test for existence of E.coli O157:H7 in excretion of mice
Finally, we testedthe excretion of all mice to ensure all of them received either E.coli O157:H7 or bacteriophage. The results from culture of excretion of mice on plates showed some plaques on overlay of E.coli O157:H7. These results indicated that there were some phages in excretion of 5 groups of mice under phage therapy. Recent studies have shown high efficacy of using phages against E.coli O157:H7. Use of E.coli O157:H7 special phages resulted in significant and log-unit reduction in E.coli O157:H7 numbers. In this way, we observed live E.coli O157:H7 from excretion of control group, so it would be expected;and special phages vs. E.coli O157:H7 from groups 1, 2, 3, 4 and 5.
Discussion
This study successfully demonstrated that special bacteriophagesreduced inoculated E.coli O157:H7 levels in animalmodel. One of the few published studies that have used animal system to evaluate the activity of phage against E.coliO157:H7 is a study by Bach et al (2009) also,E.coli O157:H7 was found to survive the animal model for over 24 h, whereas Bach et al (Ibid) reported a natural decline of E.coli O157:H7 in a continuous system over a longer incubation time (192 h). Indeed, E.coli O157:H7 in the present study were also found to be recovered in the rumen but were found to decline over time (92 h). Shedding of E.coli O157:H7 also appeared to vary among animals in the trials, but the numbers of E.coli O157:H7 shed usually decreased over time, and most animals remained E.coli O157:H7 enrichment positive through to the end time of the trials. Other researchers have reported heterogeneity of E.coli O157:H7 carriage and excretion rates among animals. The factors that contribute to the persistence of E.coli O157:H7 in animal model are unclear, but it has been suggested that the model may be a transient site for E.coli O157:H7and that the microorganism primarily colonizes at the recto-anal junction (RAJ) of the animal(Naylor et al., 2003).It was previously reported that the application of phage directly to the RAJ, as well as the addition of the phage to the drinking water, significantly reduced E.coli O157:H7 populations but did not eliminate the organism in cattle(Walker et al., 2010).It was encouraging to find that the phages in the present study were recoverable from the feces of the yearling animals, as it demonstrated that the phages can survive transit through the GITs of these animals. It may be possible in the future to incorporate the phages into protective delivery systems, such as microencapsulation, which could aid in increasing the protection of the phages from inhibition and successfully transferring higher numbers of phages to target areas within the GIT, such as the RAJ(Bach et al., 2009).So, we can say, bacteriophagetherapy has a potential for many applications, most importantly for the prevention and treatment of infections due to multiple drug resistant organisms(Merabishvili et al., 2012).45 samples from 70 were contained lytic phages. Among these, 19 samples had specific lytic activity against E.coliO157:H7. The phages in this study had isolated from wastewater. Finally it was observed that 11 samples were active against all four strains. Plaque formation and antibiogram test by overlay method and disks was detected,respectively. Five samples effect on EPEC, so had specific phages vs. bacteria, and ten samples on both ETEC and EIEC. At last, 19 samples were active against E.coli O157:H7.Among these methods, overlay used for rapid detection, but antibiogram was the best method. Capparelli et alinvestigated on lytic bacteriophages isolated from cow manure and its impact on bacteria E.coli O157:H7 in mice. Phages acquire stability in the circulatory system of mice within 38 days. After infecting mice with bacteria(1×107cfu/ml) dose, bacteria were gone from the animal within 48 hours, so about 60% of the mice survived and 40% died. The mortality will occur due to the release of Shiga toxin and rapid lysis of bacteria by phages(Capparelli et al., 2006). Tanji et al (2005) evaluated the combination effect of three bacteriophage sp15-sp21-sp22 on E.coli O157:H7 in the digestive tract. Addition of one or two phages reduced the accumulation of bacteria under anaerobic conditions. Reductionin concentration of bacteria was observed after adding phage (1×108pfu/ml). Repeat in addition of phage cocktail E.coli was effective in reducing the phage cocktail. Existence of bacteria in the gastrointestinal tract of mice was decreased when the excretion was full of phages. Repeating administration of phage (sp15-sp21-sp22) was useful for purifying E.coliO157:H7 from manure and digestive tract of mice(Tanji et al., 2005).In presence study, we repeat the processes of feeding in rats; it resulted in clearing phages from the digestive tract of mice. Also phage suspension (25.0% calcium carbonate) could maintain the pH of the stomach in favorite manner for phage therapy. The main differences of our work with Tanji et al (2005) was that we administrated phages on the same day that the bacteria enteredinto mice but theylong it two days more.In this study, to evaluate the effect of specific lytic phages isolated from sewage, an animal model was used. Six Groups in 3 repeat selected in similar conditions. The results showed that four specific lytic phage effect on E.coli O157:H7 but one group was not. In mixed group, there were not any significant differences, but in the control group, on the second day after gavage diarrhea was observed that resulted in a general weakness, loss of mobility and weakening of the mice. The results showed the effectiveness of the phage about lack or inhibition of growth of bacteria that can confirm the presence of phage effect in the treatment of E.coli O157:H7.
Conclusion
Bacteriophages are a diverse group of viruses which are easily manipulated, and therefore have potential uses in biotechnology, research, and therapeutics. It is a fact that, now, from years ago, we have reached a critical point in treating infectious diseases: new drugs are not being developed at anywhere near the pace necessary to keep ahead of the natural ability of bacteria to evolve and defend themselves against antibiotics. The result is that some of our most powerful drugs are becoming useless. Finally, it can be said that the isolated phages was highly specific for E.coli O157:H7.Several suggestions are shared in order to point a direction for overcoming common obstacles in applied phage technology. The key to successful use of phages in modern scientific, farm, food processing and clinical applications is to understand the common obstacles as well as the best practices and to develop answers that work in harmony with nature.
References
- World Health Organization. WHO global strategy for containment of antimicrobial resistance, 2001.
- W. Boucher, G. H. Talbot, J. S. Bradley, J. E. Edwards, D. Gilbert, L. B. Rice, J. Bartlett. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clinical Infectious Diseases, 2009.
CrossRef - Sheng, H. J. Knecht, I. T. Kudva, and C. J. Hovde. Application of bacteriophages to control intestinal Escherichia coli O157: H7 levels in ruminants. Applied and Environmental Microbiology, 2006; pp. 5359-5366.
CrossRef - M. Knoll and E. Mylonakis. Antibacterial bioagents based on principles of bacteriophage biology: an overview. Clinical infectious diseases, 2014; pp. 528-534.
CrossRef - Rivas, B. Coffey, O. McAuliffe, M. J. McDonnell, C. M. Burgess, A. Coffey, G. Duffy. In vivo and ex vivo evaluations of bacteriophages e11/2 and e4/1c for use in the control of Escherichia coli O157: H7. Applied and environmental microbiology, 2010, pp. 7210-7216.
CrossRef - C. Keen. Felix d’Herelle and our microbial future. Future microbiology, 2012; pp. 1337-1339.
CrossRef - Adhya and C. Merril. The road to phage therapy. Nature, 2006; pp. 754-755.
CrossRef - A. Jassim, R. G. Limoges. Natural solution to antibiotic resistance: bacteriophages ‘The Living Drugs’. World Journal of Microbiology and Biotechnology, 2014; pp. 2153-2170.
CrossRef - Weber-Dąbrowska, M. Mulczyk, and A. Górski. Bacteriophage therapy of bacterial infections: an update of our institute’s experience, 2001.
- J. Marinelli, S. Fitz-Gibbon, C. Hayes, C. Bowman, M. Inkeles, A. Loncaric, J. Kim. Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates. MBio, 2012.
CrossRef - M. La Ragione, A. Best, M. J. Woodward, A. D. Wales. Escherichia coli O157: H7 colonization in small domestic ruminants. FEMS microbiology reviews, 2009; pp. 394-410.
CrossRef - Tomat, L. Migliore, V. Aquili, A. Quiberoni, C. Balagué, C. Phage biocontrol of enteropathogenic and shiga toxin-producing Escherichia coli in meat products. Frontiers in cellular and infection microbiology, 2014.
- S. Shahrbabak, Z. Khodabandehlou, A. R. Shahverdi, M. Skurnik, H. W. Ackermann, M. Varjosalo, and Z. Sepehrizadeh. Isolation, characterization and complete genome sequence of PhaxI: a phage of Escherichia coli O157: H7. Microbiology, 2013; pp. 1629-1638.
CrossRef - Krüger and P. M. Lucchesi. Shiga toxins and stx phages: highly diverse entities. Microbiology, 2015; pp. 451-462.
- R. Wagner. Classification and nomenclature of viruses: Fifth Report of the International Committee on Taxonomy of Viruses, Archives of Virology Supplementum 2. Edited by RIB Francki, CM Fauquet, DL Knudson and F. Brown. Springer Verlag, Wien/New York 1991. 450 pages, ÖS 770, DM 110 (reduced rate for subscribers of archives of virology), 1992.
- A. Ferens, and C. J. Hovde. Escherichia coli O157: H7: animal reservoir and sources of human infection. Foodborne pathogens and disease, 2011; pp. 465-487.
CrossRef - J. Payne, M. Petrovic, R. J. Roberts, A. Paul, E. Linnane, M. Walker, G. Willshaw. Vero cytotoxin-producing Escherichia coli O157 gastroenteritis in farm visitors, North Wales. Emerging infectious diseases, 2003; pp. 526-530.
CrossRef - A. Chapman, J. Cornell, and C. Green. Infection with verocytotoxin-producing Escherichia coli O157 during a visit to an inner city open farm. Epidemiology and infection, 2000; pp. 531-536.
CrossRef - M. Thorpe, J. M. Ritchie, and D. W. K. Acheson. Enterohemorrhagic and other Shiga toxin-producing Escherichia coli. Escherichia coli: Virulence mechanisms of a versatile pathogen, 2002.
- J. Bach, T. A. McAllister, D. M. Veira, V. P. Gannon, and R. A. Holley. Effect of bacteriophage DC22 on Escherichia coli O157: H7 in an artificial rumen system (Rusitec) and inoculated sheep. Animal Research, 2003; pp. 89-101.
CrossRef - Chase-Topping, D. Gally, C. Low, L. Matthews, M. Woolhouse. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nature Reviews Microbiology, 2008; pp. 904-912.
CrossRef - W. Naylor, J. C. Low, T. E. Besser, A. Mahajan, G. J. Gunn, M. C. Pearce, M. C, D. L. Gally. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157: H7 in the bovine host. Infection and immunity, 2003; pp. 1505-1512.
CrossRef - Walker, X. Shi, M. Sanderson, J. Sargeant, and T. G. Nagaraja. Prevalence of Escherichia coli O157: H7 in gut contents of beef cattle at slaughter. Foodborne pathogens and disease, 2010; pp. 249-255.
CrossRef - J. Bach, R. P. Johnson, K. Stanford, and T. A. McAllister. Bacteriophages reduce Escherichia coli O157: H7 levels in experimentally inoculated sheep. Canadian journal of animal science, 2009; pp. 285-293.
CrossRef - Merabishvili, D. De Vos, G. Verbeken, A. M. Kropinski, D. Vandenheuvel, R. Lavigne, J. Scheres. Selection and characterization of a candidate therapeutic bacteriophage that lyses the Escherichia coli O104: H4 strain from the 2011 outbreak in Germany. PLoS One, 2012.
CrossRef - Capparelli, I. Ventimiglia, S. Roperto, D. Fenizia, and D. Iannelli. Selection of an Escherichia coli O157: H7 bacteriophage for persistence in the circulatory system of mice infected experimentally. Clinical microbiology and infection, 2006; pp. 248-253.
CrossRef - Tanji, T. Shimada, H. Fukudomi, K. Miyanaga, Y. Nakai, H. Unno. Therapeutic use of phage cocktail for controlling Escherichia coli O157: H7 in gastrointestinal tract of mice. Journal of bioscience and bioengineering, 2005; pp. 280-287.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





