How to Cite | Publication History | PlumX Article Matrix
Records on Associated Endosymbionts and Genetic Group of Bemisia tabaci (Gennadius) Feeding on Okra
Tahseen R. Hashmi1,2*, D. Dey1,Salam Rita Devi1, Ajit Varma2 and Ram Prasad2
1Division of Entomology, Indian Agricultural Research Institute, New Delhi- 110012, India.
2Amity Institute of Microbial Technology, Amity University, Uttar Pradesh- 201313, India. Corresponding Author E-mail: findtahseen@gmail.comDOI : http://dx.doi.org/10.13005/bbra/2403
ABSTRACT: Okra (Abelmoschus esculentus L.), is an important crop cultivated in Brazil, southern USA and India for its long, numerous seeded pods, used as a vegetable. A study was carried out to spot the dispersal frequency of bacterial endosymbionts and the genetic group of Bemisia tabaci feeding on okra plants, collected from Indian Agricultural Research Institute, New Delhi, India. The B. tabaci samples were examined based on mitochondrial cytochrome oxidase I sequences and settle down along with Asia II 1 population. Dispersal frequency of seven known endosymbionts namely Portiera, Rickettsia, Wolbachia, Cardinium, Fritschea, Hamiltonella and Arsenophonus were documented. The primary endosymbiont, Portiera aleyrodidarum was present in all the studied samples and a disparity was noted in the dispersal frequency of secondary endosymbionts. The statistics of irregular dispersal of secondary endosymbionts and the genetic group of B. tabaci delivers the elementary data of this notorious pest for advance studies on the control measures of this insect pest over okra plantation.
KEYWORDS: Bemisia tabaci; genetic group; vegetable; endosymbionts
Download this article as:| Copy the following to cite this article: Hashmi T. R, Dey D, Devi S. R, Varma A, Prasad R. Records on Associated Endosymbionts and Genetic Group of Bemisia tabaci (Gennadius) Feeding on Okra. Biosci Biotech Res Asia 2016;13(4). |
| Copy the following to cite this URL: Hashmi T. R, Dey D, Devi S. R, Varma A, Prasad R. Records on Associated Endosymbionts and Genetic Group of Bemisia tabaci (Gennadius) Feeding on Okra. Biosci Biotech Res Asia 2016;13(4). Available from: https://www.biotech-asia.org/?p=17439 |
Introduction
India has extensive past of vegetarianism and the core vegetable crops are brinjal, chili tomato, okra, cabbage and cauliflower etc.1, which struggles from numerous insect pests. Amongst the insect pests, sweet potato whitefly, Bemisia tabaci (Gennadius) described as pest of tobacco in 18892 is a foremost threat worldwide nourishing on vegetables, fruit crops3 and pulses from 86 botanical families4. Adults and nymphs are destructive stages and usually found resting underside of the leaves. Other than sap sucking5 and excreting honey dew6, whitefly have a role in transmission of more than 115 types of virus7 to the commercial crops amongst which 90% belong to Begomovirus genus8.
Bemisia tabaci (Gennadius) is globally known as economically vital insect pest and is polyphagous in nature. It is dispersed throughout the northern and western regions of the Indian subcontinent, and has freshly arisen as a very serious pest in okra plantation and production. Bemisia tabaci is a phloem feeder as we know that plant phloem is highly enriched with carbohydrate content and due to which it lacks essential amino acids required for the growth of the pest. Microbial community be inherent in the pest recompense the insufficiency of the scarce amino acids and nutritional content9. The only noted primary endosymbiont of whitefly is Portiera aleyrodidarum10 (Baumann, 2005), whereas the secondary endosymbionts have a number of bacteria like Wolbachia (Rickettsiales)11, Arsenophonus (Enterobacteriales)12, Cardinium (Bacteroidetes)13, Rickettsia (Rickettsiales)14, Hamiltonella (Enterobacteriales)11 and Fritschea (Chlamydiales)15.
Secondary endosymbionts have been labelled to have plentiful impression on the insects, such as heat tolerance16, resistivity to parasitoids17, skill of virus transmission18, and vulnerable to insecticides19-20. Invasion of rickettsia is stated to have enhancement in fitness substantially and female biasness in the host population21. The symbionts act as both mutualist and reproductive manipulator for the host insect, with understandable positive impact on host population increases as well as the spread of symbiont in fields.
Present study, will reconnoiter the genetic group and the dispersal frequency of endosymbionts residing in B. tabaci samples feeding okra plantation in New Delhi, India. Study will deliver elementary evidences on the dispersal frequency of endosymbionts and genetic group in this region on okra plantation and helps as a supportive data for the control measure of this pest over okra crops.
Material and Methods
Sampling and DNA Extraction
Samples of B. tabaci used in the existing examination were collected from arenas of Indian Agricultural Research Institute, New Delhi, India. Samples were kept in eppendorf tubes comprising 100% ethanol and stored at -200C till handled for genomic DNA isolation. Total of 60 individuals were handled as samples. Individually flies were cleaned twofold with sterile distilled water and whole genomic DNA was take out through DNASure Tissue Mini Kit (Nucleo- pore, Genetix) as per manufacturer’s protocol. The extracted genomic DNA of each replicate was kept at -200C.
Identification of B. tabaci Genetic Group
Molecular interpretation of B. tabaci for approval of the genetic group was impelled based on mitochondrial cytochrome oxidase I (mtCOI) sequences after PCR reaction with universal primers (Table 1). The PCR program for the reaction is specified in Table 2. The products were examined in 1.0% agarose gel comprising ethidium bromide under UV illumination after a passage of 45 minute at 80 V. Through the predicted band size (Table 1) on the gels, the products (20 µl) were sent for outsource sequencing. Records for sequences were explored using the BLAST algorithm in NCBI Gene Bank, and were aligned using BioEdit version 7.2.5 (Thompson et al., 1994). Distance was calculated using the Kimura 2-parameter model of MEGA 6.
Table 1: Primers used in PCR detection of endosymbionts and genetic group.
| Targeted gene | Primer’s Sequence (5’-› 3’) | Annealing temp. (0C)/ Product size (bp) | Reference |
| Portiera
16S rRNA |
F-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG
R- CCGTCAATTCMTTTGAGTTT |
60/ 550 | 32 |
| Cardinium
16S rRNA |
F- GCGGTGTAAAATGAGCGTG
R- ACCTMTTCTTAACTCAAGCCT |
58/ 400 | 13 |
| Rickettsia
16S rRNA |
F- GCTCAGAACGAACGCTATC
R- GAAGGAAAGCATCTCTGC |
60/ 900 | 14 |
| Hamiltonella
16S rRNA |
F- TGAGTAAAGTCTGGAATCTGG
R- AGTTCAAGACCGCAACCTC |
60/700 | 11 |
| Wolbachia
16S rRNA |
F- CGGGGGAAAAATTTATTGCT
R- AGCTGTAATACAGAAAGTAAA |
55/ 700 | 33 |
| Fritschea
23S rRNA |
F- TGGTCCAATAAGTGATGAAGAAAC
R- GCTCGCGTACCACTTTAAATGGCG |
60/ 600 | 34 |
| Arsenophonus
23S rRNA |
F- CGTTTGATGAATTCATAGTCAAA
R- GGTCCTCCAGTTAGTGTTACCCAAC |
60/ 600 | 12 |
| B. tabaci
MtCOI |
F- TTGATTTTTTGGTCATCCAGAAGT
R- TCCAATGCACTAATCTGCCATATTA |
52/ 800 | 35 |
Table 2: PCR programs for the detectionof prevalence of Primary and Secondary endosymbionts in B. tabaci.
| Endosymbionts | Pre- denaturation | Denaturation | Cycling conditions | ||
| Annealing | Extension | Cycles | |||
| Portiera | 94 0C (4 Min) | 94 0C (30 s) | 56 0C (2 Min) | 72 0C (2 Min) | 35 |
| Hamiltonella | 94 0C (4 Min) | 94 0C (30 s) | 52 0C (2 Min) | 72 0C (2 Min) | 35 |
| Wolbachia | 94 0C (4 Min) | 94 0C (30 s) | 55 0C (2 Min) | 72 0C (2 Min) | 35 |
| Arsenophonus | 94 0C (4 Min) | 94 0C (30 s) | 56 0C (2 Min) | 72 0C (2 Min) | 35 |
| Cardinium | 94 0C (4 Min) | 94 0C (30 s) | 52 0C (2 Min) | 72 0C (2 Min) | 35 |
| Rickettsia | 94 0C (4 Min) | 94 0C (30 s) | 58 0C (2 Min) | 72 0C (2 Min) | 35 |
| B. tabaci
mtCOI |
94 0C (1 Min) | 94 0C (1 Min) | 55 0C (1 Min) | 72 0C (1 Min) | 35 |
Screening of Endosymbionts
All the samples were checked autonomously for the incidence of endosymbiotic bacteria using specific primers amplifying the 16S rRNA gene for Portiera, Cardinium, Rickettsia, Wolbachia and Hamiltonella, and the 23S rRNA gene for Arsenophonus and Fritschea (Table 1). PCR reaction mixture’s concluding volume of 25µl, embraces of 12.5 µl Thermo Scientific maxima hot start PCR master mix, 8.5 µl molecular grade water, 1 µl of each forward and reverse primers and 2 µl genomic DNA. The PCR program for the endosymbionts is itemized in Table 2. The products were visualized in 1.0% agarose gel containing ethidium bromide under UV illumination after a migration of 45 minute at 80 V. With the expected band size on the gels, the products were used for outsource sequencing (Table 1). The gained sequences were allied to the available sequences in the databank using BLAST algorithm in NCBI.
Results
The population of B. tabaci collected from okra plantation was evaluated with the reference sequences from NCBI and the phylogenetic analysis confirms, the specimens from okra belongs to Asia I genetic group (Fig. 1).
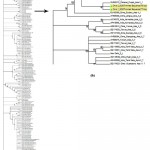 |
Figure 1: (a) Representing the phylogenetic status of B. tabaci collected from okra plantation; (b) Magnified tree |
The outcomes of the present study update the circulation frequency of seven known endosymbionts in the studied flies from okra plantation and revealed a miscellaneous spreading array. All the individuals were positive with the invasion of Portiera (Primary endosymbiont) that correspondingly measured as the positive control for the class extraction of DNA. Figure 2 is the graphical exhibition of the dispersal frequency of secondary endosymbionts in the studied B. tabaci from okra plantation. Excluding Fritschea and Hamiltonella, individuals were found verminous with rest known secondary endosymbionts unevenly. The okra collected population were found infected with Arsenophonus (30%), Rickettsia (26.66%), Wolbachia (3.33%) and Cardinium (56.66%).
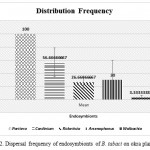 |
Figure 2: Dispersal frequency of endosymbionts of B. tabaci on okra plantation. |
Discussion
In the current study, samples were collected from grounds of Indian Agricultural Research Institute, New Delhi. This is a basic report on the genetic groups and their associated endosymbiotic microbiota investigated on one of the important host plant of B. tabaci. The examination description reveals that the specimens collected from okra belongs to most prevalent genetic group in the region i.e. Asia II 1 genetic group. The study ropes with the earlier conclusions that the range of genetic group in north and north-west India is restricted to Asia II 1, and Asia I with exceptional presence of Asia II 7 in Delhi and the occurrence of MEAM1 in some pockets of Gujrat22-24.
For the existence, passage and evolution of B. tabaci, the bacterial endosymbionts show a significant role12. The study designed on the accompanying endosymbionts of B. tabaci has been completed by many of researchers around the globe25-28 but a very limited work from India has been reported23-24, 29-30. Therefore, this study was carried out to give some extension in the evidence of associated endosymbionts of B. tabaci feeding on okra plantation in New Delhi, India.
The present study also discloses the percentage dispersal frequency of secondary endosymbionts in the flies feeding on okra plantation. Results disclosed the 100% presence of primary endosymbionts, Portiera and a dissimilarity was detected in the percentage circulation of secondary endosymbionts on the studied host plant. The flies feeding on okra belongs to Asia II 1 genetic group and it harbors Arsenophonus (30%), Rickettsia (26.66%), Wolbachia (3.33%) and Cardinium (56.66%), chains the verdict earlier reported on the host belongs to family Solanaceae and cotton22-24,29. Thus, outputs approve the association between the endosymbiotic bacterial groups and the genetic groups of B. tabaci and come to a contract with earlier works25-26,31.
The current study was emphasized in the direction of recording of the endosymbiont range associated with B. tabaci on okra plantation in New Delhi, India. The consequences stipulate, there is a lacuna present in the information of dispersal of secondary endosymbionts with respect to the host plants and genetic groups; and proposes an obligation for progressive assessments on the host wise frequency circulation of secondary endosymbionts and its term with several genetic groups of B. tabaci. A progressive and relative examination to disclose the facts regarding the role of these endosymbionts and the origin of uneven circulation, their efforts if any in the polyphagus nature of this insect pest is required for working on the control measure of this devastating insect pest.
References
- Johnson, G.I., Weinberger, K., Wu, M.H. The vegetable industry in tropical Asia: An overview of production and trade, with a focus on Thailand, Indonesia, the Philipines, Vietnam, and India [CD-ROM]. Shanhua, Taiwan: AVRDC – The world vegetable centre. 2008; 56 pp. (Explorations series; no. 1).
- Lin, K., Wu, K., Zhang, Y., Guo, Y. Overwintering and population dynamics of Bemisia tabaci biotype B in greenhouse during the spring in northern China. Crop Protec. 2007; 26:1831–1838.
CrossRef - Naveed, M., Salam, A., Saleem, M.A. Contribution of cultivated crops, vegetables, weeds and ornamental plants in harboring of Bemisia tabaci (Homoptera: Aleyrodidae) and associated parasitoids (Hymenoptera: Aphelinidae) in cotton agro-ecosystem in Pakistan. J. Pest Sci. 2007; 80:191–197.
CrossRef - Salas, J., Mendoza, O. Biology of the Sweetpotato Whitefly (Homoptera: Aleyrodidae) on Tomato. The Florida Entomol. 1995; 78(1):154–160.
CrossRef - Khan, M.R., Ghani, I.A., Khan, M.R., Ghaffar, A., Tamkeen, A. Host plant selection and oviposition behavior of whitefly Bemisia tabaci (Gennadius) in a mono and simulated polyculture crop habitat. Afr. J. Biotech. 2011; 10(8):1467–1472.
- Kakimoto, K., Inoue, H., Yamaguchi, T., Ueda, S., Honda, K., Yano, E. Host plant effect on development and reproduction of Bemisia argentifolii Bellows et Perring (B. tabaci [Gennadius] B-biotype) (Homoptera: Aleyrodidae). Appl. Entomol. Zool. 2007; 42(1):63–70.
CrossRef - Luan, J., Li, L., Varela, N., Wang, Y., Li, F., Bao, Y., Zhang, C., Liu, S., Wang, X. Global Analysis of the Transcriptional Response of Whitefly to Tomato Yellow Leaf Curl China Virus Reveals the Relationship of Coevolved Adaptations. J. Virol. 2011; 85(7):3330–3340.
CrossRef - Jones, D.R. Plant viruses transmitted by whiteflies. Eur. J. Plant Pathol. 2003; 109: 195–219.
CrossRef - Douglas, A.E. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 1998; 43: 17–27.
CrossRef - Baumann, P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 2005; 59: 155–189.
CrossRef - Zchori-Fein, E., Brown, J.K. Diversity of prokaryotes associated with Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Ann. Entomol. Soc. Am. 2002; 95(6): 711–718.
CrossRef - Thao, M.L., Baumann, P. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl. Environ. Microbiol. 2004; 70(6): 3401–3406.
CrossRef - Weeks, A.R., Velten, R., Stouthamer, R. Incidence of a new sex ratio distorting endosymbiotic bacterium among arthropods. P. Roy. Soc. Lond. B. Bio. 2003: 270(1526): 1857–1865.
- Gottlieb, Y., Ghanim, M., Chiel, E., Gerling, D., Portnoy, V., Steinberg, S., Tzuri, G., Horowitz, A.R., Belausov, E., Mozes-Daube, N. Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Appl. Environ. Microbiol. 2006; 72(5): 3646–3652.
CrossRef - Everett, K.D., Thao, M., Horn, M., Dyszynski, G.E., Baumann, P. Novel chlamydiae in whiteflies and scale insects: endosymbionts ‘Candidatus Fritsche abemisiae’ strain Falk and ‘Candidatus Fritsche aeriococci’ strain Elm. Int. J. Syst. Evol. Microbiol. 2005; 55(4): 1581–1587.
CrossRef - Brumin, M., Kontsedalov, S., Ghanim, M. Rickettsia influences thermotolerance in the whitefly Bemisia tabaci B biotype. Insect Sci. 2011; 18(1): 57–66.
CrossRef - Mahadav, A., Gerling, D., Gottlieb, Y., Czosnek, H., Ghanim, M. Parasitization by the wasp Eretmocerus mundus induces transcription of genes related to immune response and symbiotic bacteria proliferation in the whitefly Bemisia tabaci. BMC Genomics. 2008; 9: 342.
CrossRef - Gottlieb, Y., Zchori-Fein, E., Mozes-Daube, N., Kontsedalov, S., Skaljac, M., Brumin, M., Sobol, I., Czosnek, H., Vavre, F., Fleury, F., Ghanim, M. The transmission efficiency of tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. J. Virol. 2010; 84(18): 9310–9317.
CrossRef - Ghanim, M., Kontsedalov, S. Susceptibility to insecticides in the Q biotype of Bemisia tabaci is correlated with bacterial symbiont densities. Pest Manag. Sci. 2009; 65(9): 939–942.
CrossRef - Kontsedalov, S., Zchori-Fein, E., Chiel, E., Gottlieb, Y., Inbar, M., Ghanim, M. The presence of Rickettsia is associated with increased susceptibility of Bemisia tabaci (Homoptera: Aleyrodidae) to insecticides. Pest Manag. Sci. 2008; 64(8): 789–792.
CrossRef - Himler, A.G., Adachi-hagimori, T., Bergen, J.E., Kozuch, A., Kelly, S.E., Tabashnik, B.E, Chiel, E., Duckworth, V.E., Dennehy, T.J., Zchori- Fein, E., Hunter, M.S. Rapid Spread of a Bacterial Symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science. 2011; 332: 254–256.
CrossRef - Ellango, R., Singh, S.T., Rana, V.S., Gayatri Priya, N., Raina, H., Chaubey, R., Naveen, N.C., Mahmood, R., Ramamurthy, V.V., Asokan, R., Rajagopal, R. Distribution of Bemisia tabaci Genetic Groups in India. Environ Entomol. 2015; 44(4): 1258–64. doi: 10.1093/ee/nvv062.
CrossRef - Hashmi, T.R., Naveen, N.C., Dey, D., Meshram, N.M., Prasad, R. Evaluation of Endosymbionts in Different Developmental Stages of Three Population of Bemisia tabaci. Int J Innov Res Adv Stud. 2016a; 3 (10): 272–275.
- Hashmi, T.R., Dey, D., Prasad, R. Diversity of Associated Endosymbionts of Bemisia tabaci (Gennadius) on Solanaceous Host Plants in India. Indian J Sci Technol. 2016b; 9(40): 1–7.
- Chiel, E., Gottlieb, Y, Zchori-Fein, E., Mozes-Daube, N., Katzir, N., Inbar, M., Ghanim, M. Biotype-dependent secondary symbiont communities in sympatric populations of Bemisia tabaci. Bull. Entomol. Res. 2007 Aug; 97: 407– 413.
CrossRef - Gueguen, G., Vavre, F., Gnankine, O., Peterschmitt, M., Charif, D., Chiel, E., Gottlieb, Y., Ghanim, M., Zchori-Fein, E., Fleury, F. Endosymbiont metacommunities, mtDNA diversity and the evolution of the Bemisia tabaci (Hemiptera: Aleyrodidae) species complex. Mol. Ecol. 2010 October; 19: 4365– 4378.
CrossRef - Chu, D., Gao, C., De Barro, P.J., Zhang, Y., Wan, F., Khan, I. Further insights into the strange role of bacterial endosymbionts in whitefly, Bemisia tabaci: Comparison of secondary symbionts from biotypes B and Q in China. B. Entomol. Res. 2011; 101: 477–486.
CrossRef - Bing, X.L., Yang, J., Zchori-Fein, E., Xiao-We, W., Shu-Sheng, L. Characterization of a newly discovered symbiont in the whitefly Bemisia tabaci (Hemiptera: Aleyrodidae). Appl. Environ. Microbiol. 2013 Jan; 79(2): 569.
CrossRef - Singh, S.T., Priya, N.G., Kumar, J., Rana, V.S., Ellango, R., Joshi, A., Priyadarshini, G., Asokan, R., Rajagopal, R. Diversity and phylogenetic analysis of endosymbiotic bacteria from field caught Bemisia tabaci from different locations of North India based on 16S rDNA library screening. Infect. Genet. Evol. 2012 Mar; 12(2): 411–419.
CrossRef - Roopa, H.K., Rebijith, K.B., Asokan, R., Mahmood, R., Krishna Kumar, N.K. Isolation and identification of culturable bacteria from honeydew of whitefly, Bemisia tabaci (G.) (Hemiptera: Aleyrodidae). Meta Gene. 2014; 2:114–122.
CrossRef - Gnankiné, O., Mouton, L., Henri, H., Terraz, G., Houndeté, T., Martin, T., Vavre, F., Fleury, F. Distribution of Bemisia tabaci (Homoptera: Aleyrodidae) biotypes and their associated symbiotic bacteria on host plants in West Africa. Insect Conserv. Divers. 2013; 6(3): 411–421.
CrossRef - Muyzer, G., Hottentrager, S., Teske, A., Wawer, C. Denaturing gradient gel electrophoresis of PCR amplified 16s rDNA- A new molecular approach to analyze the genetic diversity of mixed microbial communities, p. 1–23. In A. D. L. Akkermans JD, van Elsas, F. J. de Bruijn (ed.), Molecular microbial ecology manual 3.4.4. Kluwer Academic Publishers. 1996.
- Heddi, A., Grenier, A.M., Khatchadourian, C., Charles, H., Nardon, P. Four intracellular genomes direct weevil biology: Nuclear, mitochondrial, principal endosymbiont, and Wolbachia. P. Natl. Acad. Sci. USA. 1999; 96: 6814–6819.
CrossRef - Thao, M.L., Baumann, L., Hess, J.M., Falk, B.W., Ng, J.C., Gullan, P.J., Baumann, P. Phylogenetic evidence for two new insect-associated chlamydia of the family Simkaniaceae. Curr. Microbiol. 2003; 47: 46–50.
CrossRef - Khasdan, V., Levin, I., Rosner, A., Levin, I., Rosner, A., Morin, S., Kontsedalov, S., Maslenin, L., Horowitz, A.R. DNA markers for identifying biotypes B and Q of Bemisia tabaci (Hemiptera: Aleyrodidae) and studying population dynamics. Bull. Entomol. Res. 2005 Dec; 95: 605–613.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





