Manuscript accepted on : 15 December 2017
Published online on: --
Dhanya Narayanan Nair and S. Padmavathy
and S. Padmavathy
Department of Bioinformatics, Nirmala College for Women, Redfields, Coimbatore, Tamil Nadu-641018, India.
Corresponding Author E-mail: dhanya.dnn@outlook.com
DOI : http://dx.doi.org/10.13005/bbra/2590
ABSTRACT: B-cell lymphocyte-2 (Bcl-2) is an antiapoptotic protein, which is an important member of Bcl-2 family. The current study involves molecular docking of six antineoplastic phytocompounds from Aloe vera (L.) Burm.f. against the protein Bcl-2. Docetaxel, a known inhibitor of Bcl-2 was used as a control in this study. All the studied phytocompounds bound within the same binding pocket as that of Docetaxel and thus can be considered as potential inhibitors of Bcl-2 protein. Among the six phytocompounds studied, AVG4 showed the best docking result, with a minimum pharmacological energy, -198.9 kcal/mol, followed by AVG6 and AVG3 as the second and third best phytocompound while AVL3 has the maximum pharmacological energy -103.8 kcal/mol. AVL3 is involved in cation-pi interactions with the Tyr9 residue of the Bcl-2 protein which is not considered while calculating pharmacological energy scoring function. Calculation of energy due to cation-pi interactions may result in the increase in total binding energy of AVL3, which may significantly increase the pharmacological energy, EPharma by approximately -8 kcal/mol, resulting in another potential anticancer phytocompound.
KEYWORDS: Aloe vera; Antiapoptotic; Anticancer; Bcl-2; Aloe vera leaf skin; AVL: AVG:Aloe vera gel; Asp: Asparagine; Asn: Asparagine; Ala: Alanine; Bcl-2: B-cell lymphoma-2;Gly: Glycine; Gln: Glutamine; His: Histidine;Ile: Isoleucine; Molecular Docking; Phytocompounds; Tyr: Tyrosine; Thr: Threonine; Trp: Tryptophan;
Download this article as:| Copy the following to cite this article: Nair D. N, Padmavathy S. Molecular Docking Studies of Phytocompounds from Aloe Vera (L.) Burm. F. Having Anticancer Property, Against an Antiapoptotic Bcl-2 Protein. Biosci Biotech Res Asia 2017;14(4). |
| Copy the following to cite this URL: Nair D. N, Padmavathy S. Molecular Docking Studies of Phytocompounds from Aloe Vera (L.) Burm. F. Having Anticancer Property, Against an Antiapoptotic Bcl-2 Protein. Biosci Biotech Res Asia 2017;14(4). Available from: https://www.biotech-asia.org/?p=28484 |
Introduction
Apoptosis means programmed cell death which is essential for normal development and tissue homeostasis. It is determined by the Bcl-2 family proteins, which has the role as both anti- and pro-apoptotic regulators. Bcl-2 family proteins need to be controlled tightly since uncontrolled apoptosis can result in degenerative conditions while too little of it can contribute to cancer and autoimmune diseases. It is now widely accepted that impairment of apoptosis results in tumor development.1-4
Bcl-2 family proteins constitute of Bcl-2, Bcl-w, Bcl-XL and Mcl-1 proteins. With regards to cancer, Bcl-2 is the most important oncoprotein with anti-apoptotic activity among the Bcl-2 family proteins.5 It is also an important marker for hematopoietic cancer like breast cancer6 and lymphoma.7 Overexpression of Bcl-2 protein is common in many forms of human cancer and has resulted in decreased susceptibility to chemotherapy and increased radio-resistance.8 High levels of Bcl-2 proteins help with a new opportunity to use it as a target for various cancers and at the same time poses a challenge to find to find good Bcl-2 inhibitors with higher susceptibility to chemotherapy and reduced radio-resistance.
Molecular docking helps in understanding the protein-ligand interactions by docking the ligand into the binding pocket. iGEMDOCK is a molecular docking software with a GUI interface, which helps in molecular docking, virtual screening, and post-screening analysis. Using iGEMDOCK helps in having a real-time docking data with the GUI interface and also helps in preprocessing of the protein and ligands.9 The present study aims to identify best potential Bcl-2 inhibitor by molecular docking studies, using iGEMDOCK, from a set of phytocompounds found to be present in Aloe vera (L.) Burm.f., predicted to have antineoplastic property through in silico
Materials and Methods
Data and Databases
The 3D structure of Homo sapiens antiapoptotic protein Bcl-2 (PDB:1G5M) was retrieved from Protein Databank (http://www.rcsb.org/).10 The binding site for the downloaded protein, 1G5M, was predicted using Discovery Studio Visualizer 3.5 Fig. 1.
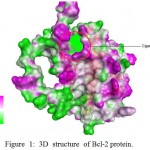 |
Figure 1: 3D structure of Bcl-2 protein.
|
The ligand binding site represented as cluster of spots in the indicated circle, within the prominent binding pocket The ligands used in this study were phytocompounds from Aloe vera (L.) Burm.f. identified using GC-MS. Through in silico studies using PASS Online tool,11 six compounds AVG3, AVG4, AVG6, AVL3, AVL5, and AVL8 Fig. 2. were found to have antineoplastic property (Table 1).
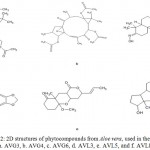 |
Figure 2: 2D structures of phytocompounds from Aloe vera, used in the study. a. AVG3, b. AVG4, c. AVG6, d. AVL3, e. AVL5, and f. AVL8
|
Table 1: Bioactivities predicted for the phytocompounds identified from Aloe vera (L.) Burm.f., using PASS Online tool
| Phytocompounds | IUPAC Name | Predicted |
| Bioactivities | ||
| AVG3 | [13,14-bis(acetyloxy)-1,5-dihydroxy- | Antiparasitic, |
| 4,12,12,15- | antineoplastic, | |
| tetramethyltetracyclo[8.5.0.0²,⁶.0¹¹,¹³]pentadeca- | antihelmintic | |
| 3,8-dien-8-yl]methyl | ||
| acetate | ||
| AVG4 | 5,8-bis(acetyloxy)-3,6,6,10,14-pentamethyl-2,9- | Antineoplastic, |
| dioxo-16- | antimicrobial | |
| oxatetracyclo[10.3.1.0¹,¹².0⁵,⁷]hexadec-10-en- | ||
| 13-yl acetate | ||
| AVG6 | [(7E)-1-(acetyloxy)-7-(hydroxyimino)-6,6,11a- | Antineoplastic, |
| trimethyl-hexadecahydro- | antiseborrheic | |
| 1H-cyclopenta[a]phenanthren-9a-yl]methyl | ||
| acetate | ||
| AVL3 | 5,7-dioxa-12- | Antineoplastic, |
| azapentacyclo[10.5.2.0¹,¹³.0²,¹⁰.0⁴,⁸]nonadeca- | antidyskinetics | |
| 2,4(8),9,16-tetraen-18-ol | ||
| AVL5 | 1-methoxy-10-methyl-5-[(1E)-prop-1-en-1-yl]- | Antineoplastic, |
| 2,6- | anti-eczematic, | |
| dioxatricyclo[8.4.0.0³,⁸]tetradecane-7,11-dione | antiinflammatory, | |
| Immunosuppressant | ||
| AVL8 | 5,14-dihydroxy-2,15- | Antineoplastic, |
| dimethyltetracyclo[8.6.0.0²,⁷.0¹¹,¹⁵]hexadecan- | general anesthetic, | |
| 16- | erythropoiesis | |
| One | stimulant, | |
| antiseborrheic, | ||
| respiratory | ||
| analeptic, alopecia | ||
| treatment, caspase 3 | ||
| stimulant, Tp53 | ||
| expression enhancer |
The structures of these ligands were constructed using ChemSketch (ACD/ ChemSketch v. 01, 2010).12 The structure of Docetaxel, a known inhibitor of the Bcl-2 protein, used in this study as a control, was retrieved from Zinc Database (http://www.zinc.docking.org/)13 with Zinc Database id: 85537053. The primary structure of the Bcl-2 protein (P10415) was retrieved from UniProt (http://www.uniprot.org).14 Various physicochemical properties of the Bcl-2 protein were computed using ProtParam (http://www.expasy.com/).15
Ligand and Protein Preparation
The 3D structure of ligands and the protein Bcl-2 were opened in Discovery Studio Visualizer 3.5 They were prepared by adding hydrogen and suitable charges, applying appropriate geometry, and removal of water molecules from the protein.
Molecular Docking with iGEMDOCK
iGEMDOCK is an interactive, GUI-based freeware docking software which can be used for virtual screening, docking, and post-screening. Accurate docking was performed for Bcl-2 protein against 7 ligands including 6 phytocompounds from Aloe vera extracts and one known inhibitor of the protein, Docetaxel. Docking settings were defined by selecting accurate docking which is very slow and precise form of docking, with population size 800, for 80 generations and number of solutions 10. Post-docking analyses for the best- docked poses were performed using the features, interaction profile, interaction analysis and interaction profile clusters.
The pharmacological scoring function for identifying the active compounds from the set of phytocompounds and known inhibitor, Docetaxel was performed. The pharmacological energy scoring function, EPharma, is given as
EPharma = EGEMDOCK + E(E)Pharma + 2E(H)Pharma + 0.5E(V)Pharma
where EGEMDOCK is the docked energy of GEMDOCK and E(E)Pharma, E(H)Pharma, and E(V)Pharma are the pharmacological scores of electrostatics, hydrogen-bonding, and VdW interactions, respectively.9
Visualization of Protein-Ligand Interactions
Protein-ligand interactions for each best-docked poses were visualized and analyzed using Discovery Studio Visualization 3.5. The protein-ligand interactions both in 2D and 3D form were obtained in image format, all in publication quality.
Results and Discussions
Bioinformatics Computation of Bcl-2 Protein
ProtParam analysis shows Bcl-2 protein is 239 amino acids long, with theoretical PI 75. Extinction coefficient indicates how much light a protein absorbs at a certain wavelength. The extinction coefficient for the Bcl-2 protein assuming all pairs of Cys form Cysteines is 1.715 and all Cys residues are reduced, it is 1.710. The estimated half-life is 30 hrs for in vitro mammalian reticulocytes, >20 hrs for in vivo yeast and >10 hrs for in vivo Escherichia coli. The instability index of the protein is computed to be 51.63. The instability index greater than 40, indicates it is an unstable protein. Since ProtParam analysis is done using the primary structure of the protein, does not necessarily designate the Bcl-2 protein as unstable in native form. Studies have shown that Bcl-2 protein is stable, and its high efficiency, compared to few other Bcl-2 protein family members, is due to its stability.16 So, it can be hypothesized that post-translational modification can be a contributor to the stability of the Bcl-2 protein. The aliphatic index of the protein 78.03. The higher aliphatic index indicates higher thermostability,17 hence Bcl-2 is a thermostable protein. The grand average of hydropathy (GRAVY) is -0.136
Molecular Docking and Post-Docking Analysis
Pharmacological and energy-based scoring of ligands
The energy EGEMDOCK is the total docked energy computed by GEMDOCK while EPharma is the pharmacological based scoring function which helps to identify the active compounds from the set of six phytocompounds from Aloe vera. In this study, Docetaxel has the lowest energy -180.9 and -239.4, computed based on EGEMDOCK and EPharma, respectively, and hence must be ranked highest, but are not ranked among the other Aloe vera phytocompounds, since it was used as control in this study, a known inhibitor of Bcl-2 protein and hence was not necessary to rank among the others. Among the phytocompounds from Aloe vera studied, AVG4 was ranked highest with the minimum energies based on EGEMDOCK and EPharma, -141.7 and -198.9 kcal/mol, respectively (Table 2).
Table 2: Pharmacological and Energy based scoring of ligand interactions with Bcl-2 protein
| Ligands | EPharma | EGEMDOCK | E_Hbond | E_Vdw | EGEMDOCK based | EPharma based |
| AVG3 | -195.0 | -135.6 | -31.6 | -104.0 | 2 | 2 |
| AVG4 | -198.9 | -141.7 | -36.5 | -105.2 | 1 | 1 |
| AVG6 | -181.1 | -128.0 | -31.7 | -96.3 | 3 | 3 |
| AVL3 | -103.8 | -85.3 | -10.9 | -74.3 | 5 | 6 |
| AVL5 | -157.6 | -96.5 | -27.9 | -68.6 | 4 | 4 |
| AVL8 | -134.2 | -84.7 | -20.6 | -64.1 | 6 | 5 |
| Docetaxel* | -239.4 | -180.9 | -50.1 | -130.8 | – | – |
*Since Docetaxel is the control drug, a known inhibitor of Bcl-2, was not ranked based on both EGEMDOCK and EPharma
The rankings of the phytocompounds based on EGEMDOCK and EPharma remain almost the same in this case except for AVL3 and AVL8, which got This is due to the improvement in screening accuracy when considered the pharmacological interactions and hence AVL8 is a better active compound compared to AVL3 as per EPharma. The developers of the software iGEMDOCK, Hsu et al,9 have mentioned that the energy scoring function EPharma is derived considering the pharmacological interactions which are often involved in the biological functions and the ligand binding.
Pharmacological Interactions
Pharmacological interactions help in understanding the binding. Docetaxel, the known inhibitor of the Bcl-2 protein, interacts with maximum residues within the binding pocket of the protein. Keeping it as reference compound, the interaction profiles for other phytocompounds are studied and are indicated in Table 3.
Table 3: The interacting amino acid residues within binding pocket of Bcl-2 protein with the ligands studied
| S. No. | Phytocompounds | Interacting amino acid residues |
| 1 | AVG3 | Ala4, Arg6, Thr7, Gly8, Tyr9, Ile189, Gln190, Trp195 |
| 2 | AVG4 | His3, Ala4, Arg6, Thr7, Gly8, Tyr9, Asn11, His186, |
| Ile189, Gln190, Trp195 | ||
| 3 | AVG6 | His3, Thr7, Gly8, Tyr9, Asn11, Ile189, Gln190, |
| Trp195, Asp196 | ||
| 4 | AVL3 | Arg6, Thr7, Gly8, Tyr9, Gln190, Gly194, Trp195, |
| Asp196 | ||
| 5 | AVL5 | Thr7, Gly8, Tyr9, Asp10, Asn11, His186, Ile189, |
| Trp195 | ||
| 6 | AVL8 | Gly8, Tyr9, Asn11, Asn182, His186, Ile189, Gln190, |
| Gly193, Gly194, Trp195 | ||
| 7 | Docetaxel | Arg6, Thr7, Gly8, Tyr9, Asp10, Asn11, Glu48, Asn182, |
| His186, Ile189, Gln190, Trp195 |
All are involved in hydrogen interactions with Tyr9, Asp10, Asn11, Trp190, and Vander Waal’s interactions with His186, Gln190, and Trp195, while the residues Tyr9, Asp10, Asn11, and Trp195 have involved in both types of pharmacological interactions viz., hydrogen and Vander Waal’s interactions. Ile189 interacts with all ligands except AVL3. None of the compounds are involved in electrostatic interactions. Thr7, Gly8, Tyr9, Asp10, Asn11, Ile189, Gln190, and Trp195 have the consensus ratio >0.5 and hence are considered hotspot residues and hence can constitute the binding pocket.
Molecular Docking Analysis
All the ligands studied i.e. all phytocompounds from Aloe vera and docetaxel are docked within the same binding pocket of Bcl-2 protein, indicated in green color Fig. 3.
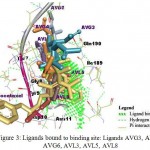 |
Figure 3: Ligands bound to binding site: Ligands AVG3, AVG4, AVG6, AVL3, AVL5, AVL8 and Docetaxel bound to the Bcl-2 protein within the same binding site are represented
|
Most of them interact with the same residues within the binding pocket. Molecular docking was performed for the complete 3D structure available (1G5M) Bcl-2 protein and not with binding pocket alone. From Fig 3, it is understood that all phytocompounds studied bind within the same region of the protein as where docetaxel binds, which means all of them have the same binding pocket. Since docetaxel is a known inhibitor of the Bcl-2 protein and all the phytocompounds binding within the same cavity, it can be confirmed all the phytocompounds are inhibitors of Bcl-2 protein. But the degree of inhibition of each phytocompound can be concluded from the binding energy.
Among the phytocompounds from Aloe vera, AVG4 Fig. 4a. and 4b. is the best ligand, as it has the minimum pharmacological energy while AVL3 Fig. 5a. and 5b. has the maximum pharmacological energy. But, though the pharmacological energy was maximum for AVL3 among all the phytocompounds studied, from Fig 5b, it can be understood that there is the presence of one each of sigma-pi of 3.8Å length and pi-pi of 5.49 Å length interactions between the benzene ring of the AVL3 and Tyr-9 residue of the Bcl-2 protein.
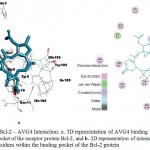 |
Figure 4: Bcl-2 – AVG4 Interaction: a. 3D representation of AVG4 binding within the binding pocket of the receptor protein Bcl-2, and b. 2D representation of interaction of AVG4 with the residues within the binding pocket of the Bcl-2 protein
|
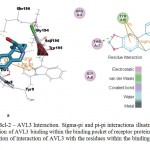 |
Figure 5: Bcl-2 – AVL3 Interaction. Sigma-pi and pi-pi interactions illustrated in both a. 3D representation of AVL3 binding within the binding pocket of receptor protein Bcl-2, and b. 2D representation of interaction of AVL3 with the residues within the binding pocket of Bcl-2 protein
|
Aromatic side chains Phenylalanine, Tyrosine and Tryptophan of a protein can contribute to cation-pi18 or sigma-pi19 or pi-pi interactions.20 The pi interactions can be considered for energy contributions along with other forms of interactions since they significantly contribute intermolecular contacts and interactions with ligands.18 Pi interaction bond length of values between 3.4 Å and 6 Å are considered favorable.21 Hence the sigma-pi and pi-pi interactions between AVL3 and Tyr9 can be well-thought-out to be Through mutational studies and molecular dynamics simulations between native and mutated protein, Singh et al,19 demonstrated that sigma-pi interactions stabilize the enzyme substrate complex of pyranose ring of xylohexose and aromatic chain of Endo-1,4-β- xylanase from Chaetomium globosum by an increase of -6.2 kcal/mol, while Boehr et al,20 proved increased pi-pi stacking energetics for enzyme substrate complex of aminoglycoside phosphotransferase –ATP/ADP by -2 kcal/mol. In our current study, as per the formula for EPharma calculation, pi interaction energetics are not considered. So if the entirety of piinteractions including one each of sigma-pi19 and pi-pi20 interactions between AVL3 and Tyr9 of Bcl-2 are considered, there can be a further significant increase in total binding energies approximately by -8 kcal/mol or even more.
Cation-pi interactions are important energetically when ligand has either a positive charge or an aromatic ring22 and are known to be important contributors of protein stability and ligand – protein interactions with most of them ranging between -3 to -6 kcal/mol while a few range between -10 to -24 kcal/mol.23 They are generally strong, a non-covalent force of interaction, and Acetylcholine, gamma-aminobutyric acid (GABA), and serotonin are examples of neurotransmitters acting as ligands for their cognate neuro-receptors that are involved in cation-pi interactions.24 Many studies have shown that increase in this energy makes them competitive with hydrogen bonds and ion pairs in drug-receptor and protein- protein interactions.25 Thus, in this case, the total binding energy of AVL3 can increase, which could further significantly increase the pharmacological energy, EPharma.
Conclusions
Among the six phytocompounds of Aloe vera studied, AVG4 has the minimum while AVL3 has maximum pharmacological energy. Though all six of the phytocompounds studied are potential inhibitors of the Bcl-2 protein, it is concluded that AVG4 is the best. Though there have been several studies that proved anticancer properties of Aloe vera extracts, this is for the first time six phytocompounds with the antineoplastic property, from vera extracts have been identified, proved through in silico studies and reported. Also, further study is needed to investigate in detail, the energy contributions due to pi interactions between AVL3 and Tyr9 residue of Bcl-2 protein to understand whether it can move to top rankings because of its change in energetics. This study further needs to be followed by in vitro and in vivo studies to prove the outcomes.
Acknowledgements
This study was performed using the resources funded by University Grants Commission, New Delhi, India, under the scheme of Innovative Programme for starting and uplifting Department of Bioinformatics at Nirmala College for Women, Coimbatore, India.
Conflict of Interest
The authors hereby declare that there is no conflict of interests regarding the publication of this research paper
References
- Cory S, Vaux D.L, Strasser A, Harris A.W, Adams J.M. Insights from Bcl-2 and Myc: Malignancy Involves Abrogation of Apoptosis as well as Sustained Proliferation. Cancer. 1999;59:1685–1692.
- Cory S, Huang D.C.S, Adams J.M. The Bcl-2 family: roles in cell survival and Oncogene. 2003;22:8590–607. DOI:10.1038/sj.onc.1207102.
CrossRef - Hanahan D, Weinberg R.A. The Hallmarks of Cancer. Cell. 2000;100: 57–70. DOI: 1016/S0092-8674(00)81683-9.
- Green R, Evan G.I. A matter of life and death. Cancer Cell. 2002; 1:19–30. DOI: 10.1016/S1535-6108(02)00024-7.
CrossRef - Deng J, Letai A. Priming BCL-2 to kill: the combination therapy of tamoxifen and ABT- 199 in ER+ breast cancer. Breast Cancer Res. 2013; 15:317. DOI:10.1186/bcr3568.
CrossRef - Dawson S.J, Makretsov N, Blows F.M, Driver K.E, Provenzano E, Le Quesne J, et al. BCL2 in breast cancer: a favourable prognostic marker across molecular subtypes and independent of adjuvant therapy received. J. Cancer. 2010;103:668–675. DOI: 10.1038/sj.bjc.6605736.
CrossRef - Davids M.S, Letai A. ABT-199: taking dead aim at BCL-2. Cancer Cell. 2013;23:139–141. DOI: 10.1016/j.ccr.2013.01.018.
CrossRef - Frenzel A, Grespi F, Chmelewskij W, Villunger A. Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis. 2009;14:584–596. DOI:10.1007/s10495-008-0300-z.Bcl2.
CrossRef - Hsu K.C, Chen Y.F, Lin S.R, Yang J.M. iGEMDOCK: a graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinformatics. 2011;12: 33. DOI:10.1186/1471-2105-12-S1-S33.
CrossRef - Rose P.W, Beran B, Bi C, Bluhm W.F, Dimitropoulos D, Goodsell D.S, et al, The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39(1):392-401. DOI: 10.1093/nar/gkq1021.
CrossRef - Languin A, Stepanchikova A, Filimonov D, Poroikov V. PASS: prediction activity spectra for biologically active substances. Bioinformatics. 2000; 16(8):747-748. DOI:10.1093/bioinformatics/16.8.747.
CrossRef - ACD/ ChemSketch, version 12.01. Advanced Chemistry Development, Inc., Toronto, ON,Canada. 2010.
- Irwin J.J, Shoichet B.K. ZINC – A Free Database of Commercially Available Compounds for Virtual Screening. J Chem Inf Model. 2005; 45:177–182. DOI: 10.1021/ci049714+.
CrossRef - The Uniprot Consortium. The Universal Protein Resource ( UniProt ). Nucleic Acids Res. 2008;36:190–195. DOI:10.1093/nar/gkm895.
CrossRef - Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins M.R, Appel R.D,Bairoch B. Protein Identification and Analysis Tools on the ExPASy Server. In: The Proteomics Protocols Handbook (Walker J.M, ed), Humana Press. 2005;571-607.
CrossRef - Rooswinkel R.W, Kooij B.V, Vries D.E, Paauwe M, Braster R, Verheij M, Borst J. Antiapoptotic potency of Bcl-2 proteins primarily relies on their stability, not binding selectivity. Blood. 2014;123:2806–2815. DOI:10.1182/blood-2013-08-519470.
CrossRef - Ikai A. Thermostability and Aliphatic Index of Globular Proteins. J. Biochem. 1980;88:1895–1898.
- Gallivan J.P, Dougherty D.A. Cation- π interactions in structural biology. Proc. Natl. Acad. Sci. 1999;96:9459–9464. DOI: 10.1073/pnas.96.17.9459.
CrossRef - Singh R.K, Tiwari M.K, Kim I.W, Chen Z, Lee J.K. Probing the Role of Sigma Interaction and Energetics in the Catalytic Efficiency of Endo-1,4-Xylanase. Applied and Environmental Microbiology. 2012;78(24): 8817–8821. DOI:10.1128/AEM.02261-12.
CrossRef - Boehr D.D, Farley A.R, Wright G.D, Cox J.R. Analysis of the Stacking Interactions between the Aminoglycoside Antibiotic Kinase APH(3)-IIIa and Its Nucleotide Ligands. Chemistry & Biology. 2002;9:1209–1217. DOI:10.1016/S1074-5521(02)00245-4.
CrossRef - Burley S.K, Petsko G.A. Amino-aromatic interactions in proteins. FEBS Lett. 1986;203(2):139-43.
CrossRef - Zacharias N, Dougherty D.A. Cation-pi interactions in ligand recognition and catalysis.Trends Pharmacol. Sci. 2002;23: 281–7. DOI: 10.1016/S0165-6147(02)02027-8.
CrossRef - Tayubi I.A, Sethumadhavan R. Nature of Cation – pi Interactions and Their Role in Structural Stability of Immunoglobulin Proteins. Biochem. 2010;75:912–918. DOI:10.1134/S000629791007014X.
CrossRef - Dougherty D.A. Cation- π Interactions Involving Aromatic Acids. J. Nutr. 2007;134(6Suppl 1):1504–1508. 137(6 Suppl 1):1504S-1508S; discussion 1516S-1517S.
- Dougherty D.A. The Cation-π Interaction. Acc. Chem. Res. 2013;46: 885–893. DOI:10.1021/ar300265y.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





