Manuscript accepted on : 20-August-2018
Published online on: 01-09-2018
Plagiarism Check: Yes
Parthasarathy M and Joel Gnanadoss J
Microbial and Environmental Biotechnology Research Unit, Department of Plant Biology and Biotechnology, Loyola College, Chennai-600034, Tamil Nadu, India.
Corresponding Author E-mail: joelgna@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2680
ABSTRACT: Streptomyces sp. LCJ12A was isolated from the soil and sediments of Pichavaram Mangrove Forest, Tamil Nadu, India. Production of protease from the Streptomyces sp. LCJ12A was carried out by using submerged fermentation. To enhance the protease production, the fermentation medium was formulated and optimized. Different carbon, nitrogen and inducer sources were used for the optimization. In that fructose, sodium nitrate and red gram husk showed greater quantity of protease production and their different concentrations were optimized in Protease production broth (PPB). Response surface methodology (RSM) was employed for the medium optimization at a low cost for the production of industrially important enzyme. The optimized values showed that fructose at 2.0 g/L enhances the yield of protease up to 120.08±2.2 U/mL, sodium nitrate at 2.0 g/L maximize the protease production up to 180.35±1.9 U/mL and red gram husk at 2.0 g/L yields 194.16±2.2 U/mL which was 1.6 times higher when compared to the unoptimized medium. Statistical optimization by using RSM predicted that 327.16 U/mL of protease enzyme can be produced. Through experimentation based on RSM, the protease yield reached up to 323.4 U/mL. When compared to unoptimized medium, the statistically optimized medium produced 3 times higher yield. As a result of the optimization studies, an increase in protease activity was reached compared to the unoptimized conditions and thus offers a new approach for industrial enzyme production.
KEYWORDS: Alkaline Protease; Inducers and Streptomyces sp. Submerged Fermentation
Download this article as:| Copy the following to cite this article: Parthasarathy M, Gnanadoss J. J. Medium Formulation and its Optimization to Enhance Protease Production by Streptomyces Sp. Isolated from Mangroves. Biosci Biotech Res Asia 2018;15(3). |
| Copy the following to cite this URL: Parthasarathy M, Gnanadoss J. J. Medium Formulation and its Optimization to Enhance Protease Production by Streptomyces Sp. Isolated from Mangroves. Biosci Biotech Res Asia 2018;15(3). Available from: https://www.biotech-asia.org/?p=30861 |
Introduction
Proteases are the most important group of the industrial enzymes which are used commercially for various industrial purposes.1,2 Proteases are enzymes capable of hydrolyzing the peptide bond in a protein molecule. Alkaline protease have wide applications in various industries such as leather, silk, detergents, pharmaceuticals and food.3,4 Physical and nutritional factors influence the protease production and therefore optimization of the factors is essential to improve the protease yield.5,6 Our society is in a necessity of developing cost-effective, environment-friendly method for synthesizing enzymes by replacing traditional carbon and nitrogen sources in various media. Fungi were also used to improve protease production through medium optimization and such proteases were also used commercially.5 Based on the nature of the amino acids or other substituents, proteases can be subdivided. Through microbial fermentation, natural and modified commercial proteases are produced with major applications in leather bating, detergents, medicine and food processing. To produce the enzyme for commercial viability, inexpensive and optimized medium are used in large scale. A number of statistical experimental designs with Response surface methodology (RSM) have been employed for optimizing enzyme production from microorganisms.7 Response surface methodology (RSM) includes regression analysis and factorial design, which is used to study the interaction and to select the optimum conditions of variables for a desirable response by evaluating the effective factors and building models.8 The aim of the present study is to optimize suitable medium and culture conditions for production of extracellular protease by Streptomyces sp. LCJ12A under submerged fermentation.
Materials and Methods
Microorganism and Culture Conditions
Streptomyces sp. LCJ12A was isolated from the soil and sediments of the Pichavaram Mangrove Forest, Tamil Nadu, India (Latitude 11º43’04.5N and Longitude 79º79’29.4E) which produced an alkaline protease in protease production broth was used in the present study.
Submerged Fermentation (Smf)
Submerged fermentation is one of the major criteria to produce protease. In the selected strain Streptomyces sp. LCJ12A growth and protease production ability was examined using different basal medium namely Modified Nutrient Glucose Broth (MNGB), Starch Casein Broth (SCB), Protease Production Broth (PPB), Gelatin Broth (GB), Malt Yeast Extract Broth (MYEB) and Glycerol Peptone Salt Broth (GPSB). Among the six different media, PPB was selected for the production, which showed the significant growth and production of protease. The medium PPB consists of the following components (g/L): glucose- 5g, peptone- 5g, yeast extract- 5g, K2HPO4– 4g, Na2HPO4– 1g, MgCl2– 0.1g, Na2CO3– 6g. 100 mL of protease production broth (PPB) was transferred to 250 mL of conical flask and autoclaved at 121°C for 15 mins. To avoid bacterial contamination, 25 µg/mL of Nalidixic acid (Himedia, India) was added and to avoid fungal contamination, 25 µg/mL of Nystatin (Himedia, India) was added to the liquid medium. A loop full of Streptomyces sp. LCJ12A culture was taken and inoculated into the conical flask under sterile condition and incubated in the rotary shaker at 120 rpm at room temperature. At the intervals of every 48 hours, 10 mL of the culture filtrate was taken from the culture medium and centrifuged at 10,000 rpm for 10 mins at room temperature. Supernatants were collected after centrifugation was used to find out the protease activity.
Protease Assay
To determine the protease activity, casein was used as a substrate. 200 µL of crude enzyme was collected from the culture broth and mixed well with 500 µL of 0.5% (w/v) casein (pH7). Then 300 µL of 0.2M phosphate buffer was added and incubated at room temperature for 10 minutes. After incubation, 1 mL of 5% (w/v) Trichloroacetic acid (TCA)9 was added to terminate the enzyme reaction and then the reaction mixture was centrifuged at 10,000 rpm for 15 minutes at room temperature to separate the unreacted substrate casein. 1 mL of supernatant was collected after centrifugation and 5 mL of 0.4M Na2CO3 was added to the collected supernatant. 1 mL of 3-fold diluted Folin Ciocalteu’s reagent (Himedia, India) was added and mixed well. Incubate the resulting solution in dark for 30 minutes at room temperature and the absorbance was measured at 660 nm in UV- Spectrophotometer (Elico, India). The protein content was estimated by following standard protocol10 using Bovine Serum Albumin (BSA) as the standard.
Medium Optimization for Protease Production under Submerged Fermentation
The influence of different factors on the protease production were carried out in liquid culture medium in separate flasks, examining one factor at a time, keeping all other variables constant. Once the optimization was done with respect to a factor, it was incorporated into the experiment for the optimization of the next factor. The effect of different carbon sources such as starch, lactose, maltose, fructose, sucrose, mannitol and glycerol and their concentrations was studied. Effect of nitrogen sources was analysed by using different nitrogen sources such as casein, urea, sodium nitrate and ammonium nitrate and their concentration ranges from 0.5 to 5.0 g/L were studied for the production of protease. High protease activity meant that the organism was capable of producing high amounts of protease in that particular factor.
Effect of Carbon Source and its Concentration on Production of Protease
The selected medium, Protease production broth (PPB) was optimized by supplementing different carbon sources such as starch, lactose, maltose, fructose, sucrose, mannitol and glycerol at 1 g/L to determine the effect of carbon source in protease production by Streptomyces sp. LCJ12A. Simultaneously, a control medium without the addition of any carbon sources was also maintained. After screening for the carbon source favoring maximum protease production and its concentration in the range of 0.5 to 5.0 g/L was studied.
Effect of Nitrogen Source and its Concentration on Production of Protease
Nitrogen sources such as casein, urea, sodium nitrate and ammonium nitrate for protease production by the Streptomyces sp. LCJ12A were studied using protease production broth (PPB) at 5 g/L. These nitrogen sources replaced the original nitrogen source available in the medium. Simultaneously, a control medium was also maintained without the addition of any nitrogen sources. After screening for the nitrogen source favoring maximum protease production, its concentration ranging from 0.5 to 5.0 g/L was evaluated.
Effect of Natural Inducers and its Concentration on Production of Protease
The effect of natural inducers on protease production by Streptomyces sp. LCJ12A was studied using protease production broth (PPB) at 5 g/L. The production medium was amended with different natural inducers such as groundnut oil cake, sesame oil cake, red gram husk, black gram husk and green gram husk at 0.5 to 5.0 g/L concentrations in separate conical flasks and the experimental set up was incubated at room temperature under shaking condition for 12 days.
Experimental Design and Production of Protease
To determine the optimal levels of three factors, fructose (X1), sodium nitrate (X2) and red gram husk (X3) was used on production of protease. By using a set of experimental design face-centered central composite design (FCCCD) was used for improving total production of protease. To analyze the experimental design, statistical software ‘Design-Expert_11.0.6’, StateEase, Inc., (Minneapolis, USA) was used. The factors fructose (X1), sodium nitrate (X2) and red gram husk (X3) in the design was studied at three different levels (-1, 0, +1). A set of 29 experiments were carried out with three factors, including six center points for a 23 FCCCD. All the three variables, fructose (X1), sodium nitrate (X2) and red gram husk (X3) were taken at a central coded value considered as zero. The three variables ranging from minimum and maximum are investigated and the full experimental plan with respect to their values in actual and coded form are given in Table 1. The average maximum protease yield was taken as the dependent variable or response (Y) after completion of experiments. By the multiple regression procedure, a second order polynomial equation was fitted to the data. This resulted in an empirical model that related the response measured to the independent variables of the experiment. The model equation of three factor system is:
Table 1: Experimental range and levels of the four independent variables used in RSM in terms of actual and coded factors.
| Range of levels | |||||||
| Variables (g/L) | Symbol | Actual | Coded | Actual | Coded | Actual | Coded |
| Fructose | X1 | 1 | -1 | 2 | 0 | 3 | +1 |
| Sodium nitrate | X2 | 1.5 | -1 | 3 | 0 | 4.5 | +1 |
| Inducers | X3 | 1 | -1 | 2 | 0 | 3 | +1 |
Y= b0+b1X1+b2X2+b3X3+b11X12+b22X22+b33X32+b12X1X2+b23X2X3+b13X1X3
Y, predicted response; b0, intercept; b1, b2, b3 linear coefficients; b11, b22, b33 squared coefficients; b12, b13, b23 interaction coefficients. To generate response surface graphs, the above model from Design Expert software was used to obtain the optimum concentration of the medium components. The complete optimized medium composition was carried out twice in flask cultivation to validate these predictions.
Comparison Study of Original and Optimized Medium for Protease Production
A comparative study was made with optimized and unoptimized medium, to prove the efficiency of the optimized medium in the protease production by Streptomyces sp. LCJ12A. 100 mL of the modified and original medium were inoculated with the respective actinomycete mycelial disc (4 mm) and incubated on an orbital shaker (120 rpm) at 30°C. The protease activity was then assayed on alternate days till the 12th day.
Results
Isolation and Identification
The strain Streptomyces sp. LCJ12A was isolated from the collected samples and identified by using 16S rRNA partial gene sequence. The 16S rRNA partial gene sequence (1347 bp) of the present strain was submitted in GenBank database and also acquired accession number as KU921108.
Effect of Carbon Sources and Their Concentration
Effect of different carbon sources were used for the protease production such as starch, lactose, maltose, fructose, sucrose, mannitol and glycerol. Fructose yields higher protease production when comparing to other carbon sources. Different fructose concentrations were also determined (0.5 – 5g/L) on the protease production by Streptomyces sp. LCJ12A in PPB was studied. The results showed that Streptomyces sp. LCJ12A exhibited higher protease production in the liquid medium containing fructose with protease activity of 46.64±1.9 U/mL at 1 g/L than other carbon sources (Table 2).
Table 2: Effect of various carbon, nitrogen and inducer sources on protease production by Streptomyces sp. LCJ12A on 8th day at 37°C under shaking (120 rpm).
| Nutrient source | Protease yield (U/ml) |
| Carbon source (1 g/L): | |
| Starch | 15.93±2.2 |
| Lactose | 16.62±1.4 |
| Maltose | 19.83±1.2 |
| Fructose | 46.64±1.9 |
| Sucrose | 33.46±1.4 |
| Mannitol | 17.68±3.0 |
| Glycerol | 19.09±1.4 |
| Nitrogen source (5 g/L): | |
| Casein | 144.37±2.8 |
| Urea | 136.23±2.2 |
| Sodium Nitrate | 159.04±2.0 |
| Ammonium nitrate | 135.02±1.8 |
| Inducers (5 g/L): | |
| Green gram husk | 149.02±1.1 |
| Black gram husk | 150.21±0.8 |
| Red gram husk | 162.02±1.9 |
| Groundnut oilcake | 133.61±1.4 |
| Sesame oil cake | 105.74±1.2 |
The results on the effect of different concentrations of fructose on the protease production by Streptomyces sp. LCJ12A is depicted in Fig. 1. Among the different concentrations tested, fructose at 2.0 g/L increased the yield of protease with 120.08±2.2 U/mL by Streptomyces sp. LCJ12A (Fig. 1). The protease production were increased at 2.0 g/L of concentration of best carbon source but declined when the concentration was increased beyond 4.0 g/L.
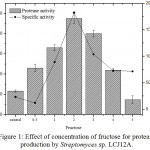 |
Figure 1: Effect of concentration of fructose for protease production by Streptomyces sp. LCJ12A.
|
Effect of Nitrogen Sources and Their Concentration
Nitrogen sources such as casein, urea, sodium nitrate and ammonium nitrate were used in protease production broth to find out the best nitrogen source for the production of protease by Streptomyces sp. LCJ12A. Sodium nitrate at 5 g/L maximizes the production of protease by Streptomyces sp. LCJ12A (Table 2). Addition of NaNO3 to the medium enhanced maximum protease production with 159.04±2.0 U/mL of protease activity by Streptomyces sp. LCJ12A. The protease activity were observed maximum on the 8th day of inoculation and declined thereafter.
The effect of different concentrations of sodium nitrate (0.5 to 5.0 g/L) by Streptomyces sp. LCJ12A was studied in PPB and the results showed that a maximum protease activity of 180.35±1.9 U/mL was observed at 2.0 g/L of sodium nitrate for Streptomyces sp. LCJ12A (Fig. 2) but declined when the concentration was increased beyond 3.0 g/L.
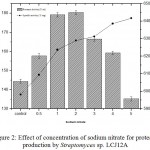 |
Figure 2: Effect of concentration of sodium nitrate for protease production by Streptomyces sp. LCJ12A.
|
Effect of Natural Inducer and Their Concentration
The effect of different natural inducers such as red gram husk, green gram husk, black gram husk, groundnut oilcake and sesame oil cake on the production of protease were studied. Addition of natural inducer (red gram husk) to the medium favored maximum protease activity of 162.02±1.9 U/mL for Streptomyces sp. LCJ12A (Table 2). Addition of red gram husk to the medium yielded higher protease production compared to the original medium.
The effect of different concentrations of red gram husk (0.5 to 5.0 g/L) by Streptomyces sp. LCJ12A was studied and results showed that a maximum protease activity of 194.16±2.2 U/mL was observed at 2.0 g/L of red gram husk by Streptomyces sp. LCJ12A (Fig. 3), the yield was higher compared to both carbon and nitrogen sources.
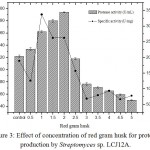 |
Figure 3: Effect of concentration of red gram husk for protease production by Streptomyces sp. LCJ12A.
|
Optimization by Response Surface Methodology
The behavior of protease production, interaction effect, main effect and squared effect (nonlinear) of three factors; fructose, sodium nitrate and red gram husk at different concentrations, were represented in three-dimensional plot (Fig. 4-6). Both fructose and sodium nitrate showed higher production of protease and the addition of inducer red gram husk also enhanced the protease production. Fructose and sodium nitrate effect of interaction was found to be highly significant (P-value of fructose and sodium nitrate = 0.01), implying that both the components were essential for production of protease. The optimal concentration of fructose and sodium nitrate were found to be 3.0 g/L and 2.0 g/L of red gram husk in response surface plot. 327.16 U/mL of maximum predicted enzyme can be produced by using optimum concentration of medium components.
 |
Figure 4: Three dimensional graph of the interaction between fructose and sodium nitrate on the protease production by Streptomyces sp. LCJ12A.
|
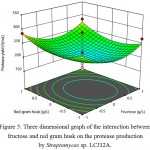 |
Figure 5: Three dimensional graph of the interaction between fructose and red gram husk on the protease production by Streptomyces sp. LCJ12A.
|
 |
Figure 6: Three dimensional graph of the interaction between sodium nitrate and red gram husk on the protease production by Streptomyces sp. LCJ12A.
|
Additional experiments in triplicate at shake flask level were performed using the optimized medium to validate the prediction of the model. These experiments yielded maximum of 323.4±0.9 U/mL enzyme (Table 3), which was three times higher than the un-optimized medium. The FCCCD result, experiments for studying the effects of three independent variables, viz., fructose, sodium nitrate and red gram husk, on production of protease are shown in Table 3 along with the observed response and mean predicted. The regression equation obtained after the analysis of variance (ANOVA) indicated the R2 value of 0.76 (a value of R2 > 0.75 indicates the aptness of the model), which ensured a satisfactory adjustment of the quadratic model to the experimental data and indicated that 90% of the variability in the response could be explained by the model. An adequate precision of 5.84 indicates an adequate signal as it measures the signal-to noise ratio. A ratio >4 is desirable. The coefficients of the regression equation were calculated using Design Expert, and the following regression equation was obtained.
Table 3: Results of FCCCD using three independent variables and six center points showing observed and predicted response.
|
Run Order |
Coded levels | Protease yield U/mL | |||
| Fructose | Sodium nitrate | Red gram husk | Observed Value | Predicted Value | |
| 1 | 1 | 1 | 0 | 312.4 | 306.29 |
| 2 | 0 | 0 | 1 | 260.3 | 254.97 |
| 3 | 0 | -1 | 0 | 250.5 | 233.79 |
| 4 | 1 | 0 | 0 | 315.3 | 327.16 |
| 5 | -1 | 1 | 0 | 296.5 | 312.55 |
| 6 | 0 | 1 | 0 | 313.2 | 311.84 |
| 7 | -1 | 0 | 1 | 286.2 | 285.28 |
| 8 | 0 | 0 | 0 | 218.4 | 201.81 |
| 9 | 0 | 1 | 0 | 285.5 | 278.91 |
| 10 | 1 | 0 | 1 | 284.3 | 273.44 |
| 11 | -1 | 0 | -1 | 281.7 | 274.54 |
| 12 | 0 | -1 | 1 | 276.3 | 295.35 |
| 13 | 0 | 1 | 1 | 284.7 | 296.53 |
| 14 | 0 | -1 | 0 | 323.4 | 311.95 |
| 15 | -1 | 0 | 0 | 296.4 | 289.83 |
| 16 | 0 | 0 | 1 | 289.4 | 275.67 |
| 17 | 0 | 0 | 0 | 165.2 | 201.81 |
| 18 | 0 | 0 | 0 | 289.5 | 201.81 |
| 19 | 0 | -1 | -1 | 263.4 | 256.90 |
| 20 | 0 | 0 | -1 | 287.5 | 314.09 |
| 21 | -1 | -1 | 0 | 283.1 | 302.00 |
| 22 | 0 | 0 | 0 | 157.4 | 201.81 |
| 23 | 0 | 0 | -1 | 164.2 | 182.31 |
| 24 | 1 | 0 | 0 | 164.2 | 189.72 |
| 25 | 0 | 1 | -1 | 314.5 | 300.74 |
| 26 | -1 | 0 | 0 | 283.6 | 263.47 |
| 27 | 0 | 0 | 0 | 178.5 | 201.81 |
| 28 | 1 | -1 | 0 | 275.1 | 271.83 |
| 29 | 1 | 0 | -1 | 267.1 | 249.96 |
Y = 201.81-9.11X1+11.25X2+8.56X3+39.89X12+56.47X22+29.11X32 +5.98X1X2 -10.67 X2X3+3.18 X1X3
Y, protease production (response); X1, fructose; X2, sodium nitrate and X3, inducer (red gram husk).
To understand the medium components interaction and the optimum concentration of each component required for maximum production of protease, three dimensional response surface curves were plotted. The coefficient of determination (R2) was 0.76. The adjusted R2, which is more suited for comparing models with different numbers of independent variables, was 0.532, depicted in Table 4. The coefficient of variance was 13.04% (Table 4). The predicted sum of squares (PRESS), which is a measure how a particular model fits each point in the design was 46171.63. This model can be used to navigate the design space.
Table 4: ANOVA for the experiments.
| ANOVA | |
| Std. Dev. | 34.47 |
| Mean | 264.42 |
| Coefficient of variance | 13.04 |
| R² | 0.7664 |
| Adjusted R² | 0.5328 |
| Predicted R² | 0.3516 |
| Adeq Precision | 5.8430 |
| PRESS | 46171.63 |
| F– Value | 3.28 |
Comparison Study of Original and Optimized Medium for Protease Production
The observed results showed that the maximum production of protease by Streptomyces sp. LCJ12A was 190.23±1.2 U/mL through conventional optimization, when the optimum values were Fructose 0.2g/100 mL, Sodium Nitrate 0.2g/100 mL and Red Gram Husk 0.2g/100 mL. Statistical optimization by RSM predicted that the yield can be reached up to 327.16 U/mL and through experimentation based on RSM, the protease yield was reached up to 323.4 U/mL, that was three times higher compared to the original medium (Table 3). Under the modified medium through conventional optimization, the observed protease activity was 190.23±1.2 U/mL whereas it was 118.41±1.1 U/mL in the original medium (Table 5). Thus, a 1.6 fold increase was obtained by conventional optimization of the factors.
Table 5: Comparison study of original and optimized medium for protease production by Streptomyces sp. LCJ12A.
| Day 2 | Day 4 | Day 6 | Day 8 | Day 10 | Day 12 | |
| Control(original medium) | 121.43±0.9 | 124.07±1.1 | 136.21±0.8 | 118.41±1.1 | 115.41±1.1 | 111.58±1.1 |
| LCJ12A(optimized medium) | 129.34±1.2 | 140.49±1.1 | 170.57±0.9 | 190.23±1.2 | 167.81±1.1 | 143.89±1.1 |
Values are mean ± standard deviation
Discussion
There is a huge need to discover new strains that produce industrially important enzymes with novel properties.11 In the current study, Streptomyces sp. LCJ12A was isolated from the mangrove environment and assessed their efficiency in the protease production, the results indicated that the selected strain has the capability to produce extracellular protease under the stagnant incubation state in the agar medium. In addition, the protease production was quantitatively determined in the liquid media under agitation condition. The results showed efficient microbial growth and potential protease production in the liquid medium. Submerged and solid-state fermentation was used for the large scale production.12,13 Submerged fermentation for the production of protease was carried out in this study. The novelty in the formulation and optimization of protease production medium for the fermentation improved the production significantly by Streptomyces sp. LCJ12A (Table 2). The most important components which are used in formulation of medium are carbon and nitrogen sources, which can influence the microbial growth and their products.14,15 Numerous researchers reported the same with various different microorganisms.16,17 Especially for the alkaline protease production, carbon and nitrogen plays a vital role in energy generation and biosynthesis. Bacteria and other microbes are most important source for the production of alkaline protease for industrial applications.18 The present study deals with the optimization of carbon source by using seven different carbon sources; starch, lactose, maltose, fructose, sucrose, mannitol and glycerol, among them fructose was found to enhance maximum protease production and minimum protease production was observed in the mannitol. Further, protease production was monitored at different concentrations of fructose (0.5 – 5g/L). That showed gradual increase of organism growth and production capability was observed while increasing the concentration of fructose at 2 g/L showed optimum production. Similarly, fructose recorded higher production of alkaline protease at the same concentration (2 g/L).19 Fructose can also be obtained from naturally occurring molasses, by utilization of inexpensive molasses as a substrate for the protease production would be a great cost effective.20
In general, microorganisms have been metabolizing nitrogen sources for their amino acids and peptides production, which are core components of an enzyme21. Commonly organic nitrogen sources provided a prominent yield of alkaline protease when compared to the inorganic nitrogen sources.22,23 The current study results revealed that prominent growth and production of Streptomyces sp. LCJ12A was observed in the inorganic nitrogen source. The current study in protease production by Streptomyces sp. LCJ12A was optimized with different inducers such as red gram husk, black gram husk, green gram husk, groundnut oilcake. Among them, red gram husk exhibits the maximum production in the liquid broth. Some have also reported that the inducer, red gram husk acts as a good substrate for production of protease.24 Additionally different concentration of red gram husk was used for the optimization, and results showed that a concentration of 2 g/L showed the higher production of protease (194.16±2.2 U/mL). By using horse gram husk as a substrate for protease production,25 reported a yield of about 80 U/ml, which is lower than the present study.
The present study deals with enhancing protease production by Streptomyces sp. LCJ12A utilizing red gram husk with optimized concentration of fructose and sodium nitrate. Thus, the optimized concentrations of both fructose and sodium nitrate along with natural inducers increased the protease production. In previous reports on the production of protease, it has been observed that the carbon and nitrogen sources are better substrates for the production of protease.26,27 Enzyme production can also be carried out by using protein-rich flours as cheaper alternative sources of carbon and nitrogen.28 On the other hand, further studies are still under investigation to fulfill the ultimate aim of maximizing the protease production in higher scales. By using response surface methodology, a number of studies have been conducted on optimization of physicochemical parameters from different organisms.29,30,31,32 Production of protease on the effects of using different carbon and nitrogen sources by Streptomyces radiopugnans_VITSD8 were examined by a full factorial design method.33 Similar work has also been done by using different carbon and nitrogen sources for the production of protease in conventional optimization, it showed that the production of the protease was high in beef extract compared to other inorganic nitrogen sources,34 whereas in the present study Streptomyces sp. LCJ12A yields maximum production in inorganic nitrogen source (sodium nitrate) given in Table 2. However, future studies on purification and characterization of Streptomyces sp. LCJ12A, would help to extent its applications in various industrial and biotechnological aspects.
Conflict of Interest
There is no conflict of interest.
Acknowldegements
We are thankful to “Times of India” and Loyola College, Chennai for providing financial support to carry out this study.
References
- Shivanand P., Jayaraman G. Production of extracellular protease from halotolerent bacterium Bacillus aguimaris strain VITP4 isolated from kumta coast. Proc Biochem. 2009;44:1088-1094.
CrossRef - Deng A., Wua J., Zhang Y., Zhang C., Wena T. Purification and characterization of a surfactant -stable high-alkaline protease Bacillus sp. B001. Bioresour Technol. 2010;101:7100-7106.
CrossRef - Benluvankar V., Priya S. E., Gnanadoss J. J. Medium Formulation and its optimization for increased protease production by Penicillium sp. LCJ228 and its potential in blood stain removal. Journal of Applied Biology & Biotechnology. 2016;4(01):020-026.
- Marchand S., Vandriesche G., Cooreviets A., Coudijzer K., Jonghe V. D., Dewettinck K. Heterogenisity of heat resistance proteases form milk Pseudomonas sp. Int J Food Microbiol. 2009;133:68-77.
CrossRef - Rani R. M., Prasad N. N., Sambasivarao K. R. S. Optimization of Cultural Conditions for the Production of Alkaline Protease from a Mutant Aspergillus flavus AS2. Asian Journal of Experimental Biological Sciences. 2012;3(3):565-576.
- Gnanadoss J. J., Devi S. K. Optimization of nutritional and culture conditions for improved protease production by Aspergillus nidulans and Aspergillus flavus. J Microbiol Biotech Food Sci. 2015;4(6):518-523.
CrossRef - Varela H., Ferrari M. D., Belobradjic L., Weyrauch R., Loperena M. L. Effect of medium composition on the production by a new Bacillus subtilis isolate of protease with promising unhairing activity. World J Microbiol Biotechnol. 1996;12:643–645.
CrossRef - Coninck D. J., Bouquelet S., Dumortier V., Duyme F., Denantes V. I. Industrial media and fermentation process for improved growth and protease production by Tetrahymena thermophila BIII. J Ind Microbiol Biotechnol. 2000;24:285–290.
CrossRef - Keay L., Wildi B. S. Proteases of the genus Bacillus, I. Neutral proteases. J Biotechnol Bioeng. 1970;12:179-212.
CrossRef - Lowry O. H., Roserbrough N. J., Farr A. L., Randall R. Protein measurement with Folin Phenol Reagent. J Biol Chem. 1951;193:265-275.
- Genckal H., Tari C. Alkaline protease production from alkaliphilic Bacillus sp. isolated from natural habitats. Enzyme Microb Tech. 2005;118:1.
- Kumar C. G, Takagi H. Microbial alkaline proteases: from a bioindustrial viewpoint. Biotechnol Adv. 1999;17(7):561-594.
CrossRef - Haki G. D., Rakshit S. K. Developments in industrially important thermostable enzymes: A review. Bioresour Technol. 2003;89(1):17-34.
CrossRef - Beg Q. K., Saxena R. K., Gupta R. De-repression and subsequent induction of protease synthesis by Bacillus mojavensis under fed-batch operations. Process Biochem. 2002;37(10):1103-1109.
CrossRef - Sharma K. M., Kumar R., Panwar S., Kumar A. Microbial alkaline proteases: optimization of production parameters and their properties. J. Genet. Eng. Biotechnol. 2017;15(1):115-126.
CrossRef - Ito E. T., Pereira G. V., Miyagui D. T., Pinotti M. H. P., Neves P. M. O. J. Production of extracellular protease by a Brazilian strain of Beauveria bassiana reactivated on coffee berry borer. Hypothenemus hampei. Braz Arch Biol Technol. 2007;50(2):1516-1531.
CrossRef - Nguyen T.T, Quyen D.T. Overproduction of an extracellular protease from Serratia sp. DT3 just using soya bean powder. World J Agric Sci. 2011;7: 29-36.
- Gupta R., Beg Q., Lorenz P . Bacterial alkaline proteases: molecular approaches and industrial applications. Appl Microbiol Biotechnol. 2002;59:15–32.
CrossRef - Abdelnasser S. S,, Ali I.A,, Al-Salamaha., Yahya B., Elbadawi., Mohamed A., El-Tayeb., Salah S., Ibrahim S. Production of extracellular alkaline protease by new halotole rant alkaliphilic Bacillus sp. NPST-AK15 isolated from hyper saline soda lakes. Electronic Journal of Biotechnology. 2015;18:236–243.
CrossRef - Hinman R. L. The changing face of the fermentation industry. Chemtech. 1994;24: 45-48.
- Niyonzima F. N., More S. Detergent-compatible proteases: microbial production, properties and stain removal analysis. Prep Biochem Biotechnol. 2015;45:233-258.
CrossRef - Kaur S. S., Chandra P. M. Characterization of thermostable alkaline proteases from Bacillus infantis SKS1 isolated from garden soil. PLoS ONE. 2015;12(11).
- Hakim A., Rumzum F. B., Iqbal A., Hossain T.E., Ahmed J., Kalam A. A. Production and partial characterization of dehairing alkaline protease from Bacillus subtilis AKAL7 and Exiguobacterium indicum AKAL11 by using organic municipal solid wastes. Heliyon. 2018;4:6.
CrossRef - Rathakrishnan and Nagarajan. Red gram husk: A potent substrate for production of Protease by Bacillus cereus in Solid-State Fermentation. International Journal of ChemTech Research. 2011;3:1526-1533.
- Shivasharanappa K ., Jayashree V., Hanchinalmath., Sundeep S.Y., Borah D., Talluri P.V. S. S. L. Optimization and production of Alkaline Proteases from Agro byproducts using a novel Trichoderma Viridiae strain VPG 12, isolated from agro soil. International Letters of Natural Sciences. 2014;14:77-84.
CrossRef - Hanlon G.W., Hodges N.A., Russel A.D. The influence of glucose, ammonium and magnesium availability on the production of protease and bacitracin by Bacillus licheniformis. J Gen Microbiol. 1982;128:845–851.
CrossRef - Kole M. M., Draper I., Gerson D. F. Production of protease by Bacillus subtilis using simultaneous control of glucose and ammonium concentrations. J Chem Technol Biotechnol. 1988;41:197–206.
CrossRef - Vidyasagar M., Prakash S. B., ramulu S. K. Optimization of culture conditions for the production of haloalkaliphilic thermostable protease from an extremely halophilic archaeon Halogeometricum sp. TSS101. Lett Appl Microbiol. 2006;43:385–391.
CrossRef - Chauhan B., Gupta R. Application of statistical experimental design for optimization of alkaline protease production from Bacillus sp. RGR-14. Process Biochem. 2004;39: 2115–22.
CrossRef - Rahman A. R., Illias M. d. R., Nawawi M. G. M., Ismail A. F., Hassan O., Kamaraduddin K. Optimization of growth medium for the production of cyclodextrin glucanotransferease from Bacillus stearothermophilus HR1 using response surface methodology. Process Biochem. 2004;39:2053–60.
CrossRef - Beg Q. K., Sahai V., Gupta R. Statistical media optimization and alkaline protease production from Bacillus mojavensis in a bioreactor. Process Biochem. 2003;39:203–9.
CrossRef - Lui H. L., Lan Y. M., Cheng Y. C. Optimal production of sulfuric acid by Thiobacillus thiooxidans using response surface methodology. Process Biochem. 2004;39:1953–61.
CrossRef - Duraikannu D., Chandrasekaran S. D. Optimization and modeling studies on the production of a new fibrinolytic protease using Streptomyces radiopugnans_VITSD8. Front. Biol. 2018:13(1):70-77.
CrossRef - Viswanathan K., Rebecca J. L, Arumugam P. Optimization of protease enzyme production by marine actinomycetes. International Journal of Pharma and Bio Sciences. 2017:8(3):188-194.

This work is licensed under a Creative Commons Attribution 4.0 International License.





