How to Cite | Publication History | PlumX Article Matrix
Nadiah Al-Sulami1, Ahmed Atef1, Mohammed Al-Matary1, Sherif Edris1,2,3, Khalid M. Al-Ghamdi1, Hassan S. Al-Zahrani1 and Ahmed Bahieldin*1,2
1Department of Biological Sciences, Faculty of Science, King Abdulaziz University (KAU), P.O. Box 80141, Jeddah 21589, Saudi Arabia.
2Department of Genetics, Faculty of Agriculture, Ain Shams University, Cairo, Egypt.
3Princess Al-Jawhara Al-Brahim Centre of Excellence in Research of Hereditary Disorders (PACER-HD), Faculty of Medicine, King Abdulaziz University (KAU), Jeddah, Saudi Arabia.
Corresponding Author E-mail: abmahmed@kau.edu.sa
DOI : http://dx.doi.org/10.13005/bbra/2747
ABSTRACT: This study aimed at studying differential presence of enzymes metabolites via KEGG analysis of trasncriptomes of the wild plant species senna (Cassia angustifolia Vahl.) due to watering. Senna is a shrub of the family Caesalpiniaceae with important applications in pharmaceuticals. Firstly, RNA-Seq datasets were produced by next-generation sequencing (NGS) of Illumina Miseq of leaf (day 1) in order to detect the influence of watering at day 2. Samples were harvested at three time points (e.g., dawn, midday and dusk) of the two days. de novo assembled datasets and number of annotated genes exceeded 2000 genes. As cluster analysis of gene expression almost showed no discrete differences at the transcriptome level due to watering within time points of dawn and dusk, the study focused mainly on those of the midday across the two days. KEGG analysis for genes whose differential expression between the two days was ≥5 FC resulted in a number of enzymes that were found repressed due to watering, thus likely participate in the molecular mechanisms utilized by the organism to adapt to the long-lasting drought stress. The recovered regulated metabolites and enzymes included abscisic acid (ABA) receptor PYL4 and PYL9, auxin response factor (ARF) 5 and 15, ARF (or Aux/IAA) proteins IAA7 and IAA14, indole-3-pyruvate (or flavin) monooxygenase, phosphoinositide phosphatase SAC1 and SAC6, pre-mRNA splicing factors 8, 8A, 19, 40A and ISY1, and serine/arginine-rich splicing regultors SCL33, RS31 and RS34. The two pathways tryptophan metabolism and plant hormone signal transduction likely crosstalk in senna (C. angustifolia) towards the maintenance of normal growth under adverse condition. Many other regulated metabolites and proteins in senna (C. angustifolia) including brassinosteroid, heat shock protein 95s, ATPase, several protein kinases such as mitogen-activated protein kinase (MAPK) and cytochrome c oxidase. Other enzymes include phospholipase C2 and allene oxide cyclase as well as isochorismate pathway were also regulated in senna (C. angustifolia). In conclusion, we think that we have scoped the light on the possible regulated metabolites under drought stress that might confer drought stress tolerance in the wild plant senna (C. angustifolia).
KEYWORDS: C. angustifolia; Isochorismate; Monooxygenase; Phosphoinositide
Download this article as:| Copy the following to cite this article: Al-Sulami N, Atef A, Al-Matary M, Edris S, Al-Ghamdi K. M, Al-Zahrani H. S. Bahieldin A. Molecular Analysis of Enzymes and Metabolites Regulated Under Drought Stress in the Wild Plant Senna (Cassia Angustifolia). Biosci Biotech Res Asia 2019;16(2). |
| Copy the following to cite this URL: Al-Sulami N, Atef A, Al-Matary M, Edris S, Al-Ghamdi K. M, Al-Zahrani H. S. Bahieldin A. Molecular Analysis of Enzymes and Metabolites Regulated Under Drought Stress in the Wild Plant Senna (Cassia Angustifolia). Biosci Biotech Res Asia 2019;16(2). Available from: https://bit.ly/2wfuvbP |
Introduction
Caesalpiniaceae is a large family with several pharmaceutical applications as being mainly used as a laxative and to relieve constipation. Cassia angustifolia, formally Senna angustifolia (2n = 28) is known for its important applications in pharmaceuticals. C. angustifolia is a wild medicinal drought-tolerant shrub (Ayoub 1977, Khalid et al. 2012) with many cathartic properties (Lemli 1986, Folkard 1995, Hammouda et al. 2005). Drought stress-related genes have been studied at the trancriptomic level in senna (C. angustifolia) (Mehta et al. 2017) as well as in many other plant species such as parsley (Li et al. 2014a,b), bean (Hiz et al. 2014), chrysanthemum (Xu et al. 2013), tall fescue (Hu et al. 2014), and grapevine (Rocheta et al. 2014). The lucine-rich repeat kinase family was recently found in senna (C. angustifolia) as the most abundant group of protein kinases under drought stress in addition to several families of transcription factors (e.g., bHLH, and bZIP, etc.) (Mehta et al. 2017). Previous studies on senna (C. angustifolia) deciphered some physiological, morphological and molecular mechanisms allowed the plant to tolerate drought stress (Ratnayaka and Kincaid 2005, Mehta et al. 2017).
Availability of water is a major obstacle for agricultural productivity. Wild plants growing in severe arid climates provide tools for studying plant response to extreme drought conditions. Generally speaking, drought, salt and heat stresses have large impacts on plant growth and productivity. Other abiotic deleterious stresses nowadays include increased chemicals and pollutants. Besides, it is likely that plants are exposed to more than one type of stress at a time. In particular, drought stress is a major threat in at least 26 % of world’s arable land (Blum 1988). The effects of drought include delayed or stunted growth as well as impaired physiological processes such as photosynthesis, respiration, and mineral exchange (Do et al. 2013). Therefore, it is crucial to get a better understanding the molecular and physiological impacts of drought stress in order to find solutions to help the plant cope with or at least lower the influence of this stress for the sake of maintaining crop productivity and possibly cultivate more crops in arid lands to mitigate global food crisis.
Plants are adapted to drought either by avoidance or tolerance, as the two main strategies, by which a crop plant can maintain yield components and minimize the loss due to the stress. Avoidance mechanisms include the occurrence of several morphological changes, such as stomatal closure and reduced leaf area to reduce respiration, as well as enlarging root systems in order to gain more water with the same intensity of cultivation (Levitt 1980, Budak et al. 2013, Rama Reddy et al. 2014). Alternatively, drought tolerance is a subject of intense research as it mainly occurs due to several physiological and molecular mechanisms that help the plant to adapt with the osmotic pressure due to the shortage of water (Bartels and Sunkars 2005). Tolerance mechanisms were proven to be genetically-dependent as different plant species have different strategies to cope with the problem. These strategies are supported by complex metabolic pathways that should link together and cross-talk in order to produce osmolytes and protein chaperons to secure the cell from the stress and avoid denaturation or damage of important compounds in the cell (Yamaguchi-Shinozaki and Shinozaki 2006, Kantar et al. 2011, Shanker et al. 2014). There are several abiotic stress-related enzymes like glutathione reductase, catalase, superoxide dismutase considered as biomarkers for drought stress tolerance (Khammari et al. 2012).
The present study aim at studying drought-related dynamics of leaf transcriptome in senna (C. angustifolia) to detect the crosstalking pathways possibly associated with drought stress tolerance to add to our understanding of the molecular mechanisms underlying drought stress tolerance in wild plants.
Material and Methods
Plant material sampling and watering regime
The field experiment of sample treatment and harvesting was conducted for senna (C. angustifolia) shrubs grown near Jeddah, Saudi Arabia. Three plants of equal size were selected in which leaf samples were harvested in two consecutive days at three time point of the day (1 h post-dawn, midday and 1 h post-dusk). At dawn of the day 2, plants were watered (25 liters dH2O) and leaf samples were harvest at the same three time points.
RNA-Seq and KEGG Analyses
Total RNA was extracted from three similar-sized (10 mm2) leaf discs per plant of Cassia angustifolia, then shipped to Beijing Genome Institute (BGI), China, for next-generation sequencing. Recovered RNA-Seq datasets were de novo assembled using the Trinity RNA-Seq Assembly package (r2013-02-25) with optimized parameters and K-mer size set to 25 (Zhang et al. 2015). Differential expression and cluster analysis were done by EdgeR (version 3.0.0, R version 2.1.5). All predicted CDS were annotated against protein database in order to assign putative function of the transcriptome after translation into protein. To identify the biological pathways with enzymes that differentiate at midday samples, the detected genes were mapped to reference canonical pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.ad.jp/kegg/). RNA-Seq datasets of Cassia angustifolia were validated via qRT-PCR (data provided upon request).
Results and Discussion
Preliminary Data Analysis
Sequence assembly resulted in a number of ˃5000 regulated genes due to watering and the number of annotated genes ˃2000 genes. The term enrichment in this study refers to the increase of a given enzyme or metabolite in the second day due to watering, while suppression indicates that the intensity of the enzyme or metabolite was reduced due to watering. In other words, enzyme or metabolite enrichment indicates that the encoding genes were highly expressed after watering, while repression indicates that the expression of the encoding drought-related genes is abolished as it is no longer required after watering. GO classification indicated that the subcategory “response to stimulus” is repressed as the stress in the second day is completely relieved. We expected that several biological processes of this subgroup can confer tolerance to drought stress.
KEGG Analysis
In order to study the enzymes in selected biological pathways of Cassia angustifolia whose genes encoding them are highly (≥5 FC) regulated due to watering, we mapped the detected genes to reference canonical pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.ad.jp/kegg/). Drought tolerance is a multigenic process with different metabolic pathways affecting plant growth (Mehta et al. 2017). Pathways that were either enriched or repressed due to watering were examined (Table S1). Of which, we selected the pathways that might have emphasis on plant response to abiotic stress (Table 1). In addition, we have investigated few other pathways whose enzymes were suppressed due to watering, while showed no enzymes that were enriched due to the stress. When applying the criteria of ≥5 FC gene regulation in KEGG analysis, a number of five groups of pathways including 14 subgroups with 22 pathways were selected. These groups are “metabolism”, “Genetic Information Processing”, “Environmental Information Processing”, “Cellular Processes” and “Organismal Systems”. Of which, 10 pathways showed no enrichment in their enzymes due to watering, rather they were repressed due to watering. This indicates that these pathways likely participate in the molecular mechanisms utilized by the organism to cope with the stress, whose enrichment is not required when water becomes available. These pathways are “Biosynthesis of siderophore group nonribosomal peptides”, “Spliceosome”, “VEGF signaling pathway”, “Jak-STAT signaling pathway”, “Autophagy – other”, “Tight junction”, “Longevity regulating pathway”, “Longevity regulating pathway – multiple species”, “Circadian rhythm” and “Thermogenesis” (Table 1). Enzymes or metabolites of the enriched and suppressed pathways are shown in Tables 2 and 3, respectively. The number of activated enzymes due to watering was 43 enzymes, while the number was 107 for the suppressed enzymes due to watering (Tables 2 and 3).
Table 1: Selected pathways of C. angustifolia with enzymes encoded by genes highly (≥5 FC) regulated due to the watering. Green box = pathway enriched due to watering, red box = pathway suppressed due to watering, blue box = regulated TOR-dependent pathway.
| Group | Subgroup | Pathway ID | Pathway | Enriched | suppressed |
| 1. Metabolism | 1.1 Carbohydrate metabolism | 562 | Inositol phosphate metabolism | ||
| 1.3 Lipid metabolism | 592 | alpha-Linolenic acid metabolism | |||
| 1.5 Amino acid metabolism | 380 | Tryptophan metabolism | |||
| 400 | Phenylalanine, tyrosine and tryptophan biosynthesis | ||||
| 1.9 Metabolism of terpenoids and polyketides | 1053 | Biosynthesis of siderophore group nonribosomal peptides | |||
| 2. Genetic information Processing | 2.1 Transcription | 3040 | Spliceosome | ||
| 2.3 Folding, sorting and degradation | 4141 | Protein processing in endoplasmic reticulum | |||
| 3. Environmental information processing | 3.2 Signal transduction | 4012 | ErbB signaling pathway | ||
| 4016 | MAPK signaling pathway – plant | ||||
| 4072 | Phospholipase D signaling pathway | ||||
| 4075 | Plant hormone signal transduction | ||||
| 4150 | mTOR signaling pathway | ||||
| 4151 | PI3K-Akt signaling pathway | ||||
| 4370 | VEGF signaling pathway | ||||
| 4630 | Jak-STAT signaling pathway | ||||
| 4. Cellular processes | 4.1 Transport and catabolism | 4136 | Autophagy – other | ||
| 4.2 Cell growth and death | 4218 | Cellular senescence | |||
| 4.3 Cellular community – eukaryotes | 4530 | Tight junction | |||
| 5. Organismal systems | 5.9 Aging | 4211 | Longevity regulating pathway | ||
| 4213 | Longevity regulating pathway – multiple species | ||||
| 5.10 Environmental adaptation | 4710 | Circadian rhythm | |||
| 4714 | Thermogenesis |
Table 2: Selected pathways of C. angustifolia with enzymes encoded by genes highly (≥5 FC) enriched at midday due to the watering. N = before watering, NR = after watering.
| No. | Pathway/Enzyme | Time point | |||||
| Before watering | After watering | ||||||
| 0380 Tryptophan metabolism | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 1 | Probable acetyl-CoA acetyltransferase cytosolic 2 | -0.24212 | -0.24212 | -0.24212 | 2.0615160 | 1.8123826 | 1.789422 |
| 2 | Amidase 1 | -0.64225 | -0.66793 | -0.67082 | 4.195597 | 2.802451 | 3.00827 |
| 3 | Aldehyde dehydrogenase family 7 member A1 | -0.50968 | -0.64755 | -0.38832 | 1.789422 | 2.655908 | 2.62422 |
| 0400 Phenylalanine, tyrosine and tryptophan biosynthesis (2) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 4 | Arogenate dehydratase/prephenate dehydratase 2- chloroplastic | -0.48907 | -0.48907 | -0.48907 | 2.473292 | 2.290608 | 2.324863 |
| 0562 Inositol phosphate metabolism (3) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 5 | Inositol oxygenase 4 | -0.97716 | -1.25336 | -1.25336 | 2.448629 | 3.041257 | 2.848035 |
| 6 | Inositol-tetrakisphosphate 1-kinase 3 | -0.09205 | 0.007279 | -0.27748 | 3.901911 | 3.545024 | 3.259165 |
| 7 | Triosephosphate isomerase cytosolic | -0.50236 | -0.14356 | 0.089004 | 3.068392 | 2.946959 | 3.529814 |
| 0592 alpha-Linolenic acid metabolism (1) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 8 | Glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a | -0.08301 | -0.29871 | -0.4084 | 2.058619 | 2.266513 | 0.83222 |
| 4012 ErbB signaling pathway (2) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 9 | Shaggy-related protein kinase kappa | -0.58381 | -0.58381 | -0.58236 | 2.858208 | 2.870764 | 3.025239 |
| 10 | Serine/threonine-protein kinase TOR | -0.62635 | -0.70496 | -0.48959 | 3.435905 | 3.497071 | 3.2715 |
| 4016 MAPK signaling pathway – plant (2) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 11 | Ethylene-insensitive protein 2 | -0.43202 | -0.43202 | -0.43202 | 1.314289 | 2.52031 | 2.397217 |
| 12 | Protein ETHYLENE INSENSITIVE 3 | -0.60329 | -1.40404 | -1.40404 | 4.847172 | 4.881385 | 4.470712 |
| 4072 Phospholipase D signaling pathway (5) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 13 | Phospholipase D zeta 1 | -0.17576 | -0.17576 | -0.17576 | 1.624987 | 1.187133 | 1.17576 |
| 14 | Dynamin-2B | -0.57567 | -0.57279 | -0.28525 | 1.046323 | 1.758612 | 2.629566 |
| 15 | Serine/threonine-protein kinase TOR | -0.60295 | -0.60295 | -0.60295 | 3.150976 | 3.324852 | 2.568417 |
| 16 | ADP-ribosylation factor 1 | -0.65602 | -0.65602 | -0.65602 | 3.214932 | 2.680402 | 3.423353 |
| 17 | ADP-ribosylation factor 2 | -1.07784 | -1.07784 | -1.07784 | 4.767723 | 4.261117 | 3.38493 |
| 18 | Ankyrin repeat- PH and SEC7 domain containing protein secG | -0.5132 | -0.5132 | -0.5132 | 1.569848 | 1.580703 | 0.971201 |
| 4075 Plant hormone signal transduction (5) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 19 | Auxin response factor 1 | -0.51286 | -0.51286 | -0.51286 | 3.266296 | 1.894222 | 2.251403 |
| 20 | Histidine-containing phosphotransfer protein 1 | -0.37534 | -0.37534 | -0.37534 | -0.37534 | 2.328646 | 1.703269 |
| 21 | Gibberellin receptor GID1B | -1.05549 | -1.14079 | -1.36132 | 3.321495 | 2.686399 | 2.662871 |
| 22 | Ethylene-insensitive protein 2 | -0.43202 | -0.43202 | -0.43202 | 1.314289 | 2.52031 | 2.397217 |
| 4141 Protein processing in endoplasmic reticulum (9) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 23 | Cullin-1 | -0.34984 | -0.24015 | -0.34984 | 0.731499 | 1.660223 | 1.86925 |
| 24 | Heat shock cognate protein 80 | -0.29447 | -0.23504 | -0.15431 | 2.723259 | 2.478121 | 1.853568 |
| 25 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit STT3A | -0.24265 | -0.12895 | -0.24265 | 1.843793 | 1.209944 | 1.24265 |
| 26 | Calreticulin | -0.86438 | -0.86438 | -0.86438 | 3.981055 | 4.049367 | 3.634441 |
| 27 | Heat shock protein 90-5- chloroplastic | -0.88796 | -0.34992 | -0.3892 | 4.341242 | 3.589975 | 2.973597 |
| 28 | Alpha-mannosidase I MNS4 | -0.54941 | -0.54941 | 0.088435 | 1.76006 | 3.3388 | 2.170433 |
| 29 | Protein transport protein Sec61 subunit alpha | -0.10453 | -0.3406 | -0.45163 | 1.002018 | 1.709612 | 2.032249 |
| 30 | Derlin-1 | -0.91968 | -0.5126 | 0.469884 | 3.480923 | 3.095565 | 2.875045 |
| 31 | Plant UBX domain-containing protein 4 | -0.44073 | -0.25043 | -0.44073 | 1.155248 | 2.939165 | 2.130943 |
| 4150 mTOR signaling pathway (4) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 32 | V-type proton ATPase catalytic subunit A | -0.33156 | -0.33156 | -0.33156 | 0.813791 | 2.564324 | 1.53317 |
| 33 | V-type proton ATPase subunit F | -0.88151 | -0.88151 | -0.88151 | 4.827068 | 4.137752 | 4.25776 |
| 34 | Shaggy-related protein kinase kappa | -0.76969 | -0.76969 | 0.29795 | 0.655233 | 1.469709 | 0.984771 |
| 35 | Serine/threonine-protein kinase TOR | -0.60295 | -0.60295 | -0.60295 | 3.150976 | 3.324852 | 2.568417 |
| 4151 PI3K-Akt signaling pathway (5) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 36 | Shaggy-related protein kinase kappa | -0.76969 | -0.76969 | 0.29795 | 0.655233 | 1.469709 | 0.984771 |
| 37 | Serine/threonine-protein phosphatase regulatory subunit A | -0.59834 | -0.63208 | -0.97463 | 2.142898 | 2.687689 | 2.046494 |
| 38 | Heat shock cognate protein 80 | -0.29447 | -0.23504 | -0.15431 | 2.723259 | 2.478121 | 1.853568 |
| 39 | Serine/threonine-protein kinase TOR | -0.60295 | -0.60295 | -0.60295 | 3.150976 | 3.324852 | 2.568417 |
| 40 | Heat shock protein 90-5- chloroplastic | -0.88796 | -0.34992 | -0.3892 | 4.341242 | 3.589975 | 2.973597 |
| 4213 Longevity regulating pathway – multiple species (2) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 41 | SNF1-related protein kinase regulatory subunit beta-2 | -0.20111 | -0.39645 | -0.39645 | -0.39645 | 2.435828 | 2.398065 |
| 42 | Serine/threonine-protein kinase TOR | -0.60295 | -0.60295 | -0.60295 | 3.150976 | 3.324852 | 2.568417 |
| 4218 Cellular senescence (1) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 43 | Serine/threonine-protein kinase TOR | -0.60295 | -0.60295 | -0.60295 | 3.150976 | 3.324852 | 2.568417 |
Drought stress triggers several plant responses at the gene expression levels, and likely result in the accumulation of secondary metabolites or osmolytes that help the plant stand the stress (Ramchandra Reddy et al. 2004, Ergen et al. 2009). In the present study, a large number of enzymes were found repressed due to watering, thus likely participate in the molecular mechanisms utilized to adapt to the long-lasting drought stress. Enrichment of these enzymes is not required when water becomes available, while re-enriched when land became dry again. KEGG analysis indicated that several gene families are involved as a safeguard against drought stress. Of these gene families, abscisic acid (ABA) receptor PYL seems to be required under drought stress for ABA-mediated responses such as stomatal closure (Hao et al. 2011). Suppression of two types of this receptor, namely PYL4 and PYL9 in senna (C. angustifolia) (Table 3, Figure 1), indicates that they participate in morphological changes as a mechanism of avoidance tolerance. PYL4 was reported to improved ABA-dependent inhibition of PP2CA and can modulate inhibition even in the absence of ABA (Pizzio et al. 2013). PP2CA plays a critical role to regulate both seed and vegetative responses to ABA and regulates stomatal aperture through interaction with the anion channel SLOW ANION CHANNEL1 (SLAC1) and the kinase OPEN STOMATA1 (Kuhn et al. 2006, Yoshida et al. 2006, Lee et al. 2009). In addition, ABA receptor PYL9 was proven to promote drought resistance and leaf senescence in Arabidopsis and rice (Zhao et al. 2016).
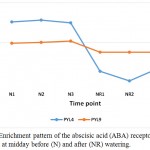 |
Figure 1: Enrichment pattern of the abscisic acid (ABA) receptors PYL4 and PYL9 at midday before (N) and after (NR) watering.
|
Auxin response factor (ARF) 5 and 15 are among the mechanisms of senna (C. angustifolia) to tolerate drought stress (Table 3, Figure 2). ARF are potential mediators of biotic and abiotic stress responses in plant (Bouzroud et al. 2018). ARF5 gene was proven to increase the contents of carotenoids and enhance the tolerance to both salt and drought when expressed in Arabidopsis (Kang et al. 2018), while ARF15 regulates cell division activity during early tomato fruit development, thus, provides important influence on plant growth under drought stress (DeJong et al. 2015). ARF5 gene also act as an activator of auxin-responsive gene expression in Arabidopsis (Remington et al. 2004). This enzyme allows the activation of ARF, derepresses downstream auxin responsive pathways, thus mediates plant growth and development (Gray et al. 2001). Also, two ARF (or Aux/IAA) proteins, namely IAA7 and IAA14, exist in stressed plant only (Table 3, Figure 3).
Table 3: Selected pathways of C. angustifolia with enzymes encoded by drought-related genes highly (≥5 FC) suppressed due to the watering. N = before watering, NR = after watering.
| No. | Pathway/Enzyme | Time point | |||||
| Before watering | After watering | ||||||
| 00380 Tryptophan metabolism (2) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 1 | Aldehyde dehydrogenase family 3 member F1 | 1.587468 | 1.287719 | -0.50763 | -1.456226318 | -1.456226318 | -1.456226318 |
| 2 | Probable indole-3-pyruvate monooxygenase YUCCA10 | 2.227227 | 2.164615 | 1.962111 | -4.308717014 | -4.308717014 | -4.308717014 |
| 00400 Phenylalanine, tyrosine and tryptophan biosynthesis (4) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 3 | Probable aminotransferase TAT2 | 0.34544 | 0.16178 | 0.060001 | -0.918195045 | -0.918195045 | -0.918195045 |
| 4 | Anthranilate synthase alpha subunit 2- chloroplastic | 0.685204 | 0.618131 | 0.077341 | -1.623098806 | -1.623098806 | -1.623098806 |
| 5 | 3-dehydroquinate synthase | 0.059773 | 0.131979 | 0.080449 | -0.258688436 | -0.258688436 | -0.258688436 |
| 6 | Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase- chloroplastic | 0.339229 | 0.373643 | -0.09642 | -2.534977735 | -2.534977735 | -2.534977735 |
| 00562 Inositol phosphate metabolism (6) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 7 | Putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1C | -0.18249 | 0.261804 | -0.48236 | -0.696482365 | -0.696482365 | -0.696482365 |
| 8 | Inositol-3-phosphate synthase | -1.33722 | -1.07768 | -2.64815 | -2.648146652 | -2.648146652 | -2.648146652 |
| 9 | Phosphoinositide phospholipase C 2 | 0.044158 | 0.002828 | -0.23448 | -3.234123788 | -3.234123788 | -2.779947895 |
| 10 | SAL1 phosphatase | -0.21719 | 0.201112 | -0.62427 | -0.624266235 | -0.624266235 | -0.624266235 |
| 11 | Phosphoinositide phosphatase SAC6 | -0.22023 | 0.207125 | 0.361932 | -1.00264063 | -1.00264063 | -1.00264063 |
| 12 | Phosphoinositide phosphatase SAC1 | -0.28007 | -0.00855 | -0.60254 | -0.689320217 | -0.689320217 | -0.689320217 |
| 00592 alpha-Linolenic acid metabolism (4) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 13 | Peroxisomal acyl-coenzyme A oxidase 1 | 0.420476 | 0.332021 | 0.490144 | -0.580245492 | -0.580245492 | -0.580245492 |
| 14 | 3-ketoacyl-CoA thiolase 2- peroxisomal | 2.497536 | 0.943436 | 0.033103 | -1.311157654 | -1.223016057 | -1.160597977 |
| 15 | Allene oxide cyclase- chloroplastic | 0.982792 | 0.939056 | 0.797635 | -4.8274607 | -4.747141535 | -4.75890803 |
| 16 | Fatty acid hydroperoxide lyase- chloroplastic | 0.794242 | 0.693469 | 0.69914 | -3.482149218 | -4.223724066 | -4.223724066 |
| 01053 Biosynthesis of siderophore group nonribosomal peptides (1) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 17 | Isochorismate synthase- chloroplastic | 0.246862 | 0.101321 | 0.281063 | -0.416710774 | -0.416710774 | -0.416710774 |
| 03040 Spliceosome (21) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 18 | Pre-mRNA-processing factor 19 homolog 2 | 0.60534 | 1.088448 | 1.057733 | -1.29615449 | -1.29615449 | -1.29615449 |
| 19 | Small nuclear ribonucleoprotein SmD1b | 1.055236 | 0.646652 | -0.28834 | -1.809891101 | -1.809891101 | -1.809891101 |
| 20 | U1 small nuclear ribonucleoprotein 70 kDa | 0.810862 | 1.303077 | -1.39203 | -1.119405074 | -1.392025529 | -1.392025529 |
| 21 | Probable small nuclear ribonucleoprotein F | 1.16508 | 0.276073 | -0.15363 | -2.104102705 | -2.104102705 | -2.104102705 |
| 22 | Sm-like protein LSM7 | 1.206399 | 0.436801 | 0.56956 | -2.583434918 | -2.583434918 | -2.583434918 |
| 23 | DEAD-box ATP-dependent RNA helicase 42 | 0.343033 | 0.826515 | 0.910273 | -3.419209265 | -3.419209265 | -3.419209265 |
| 24 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DEAH1 | 0.070534 | 0.243749 | -0.15351 | -0.153506306 | -0.153506306 | -0.153506306 |
| 25 | Pre-mRNA-processing protein 40A | 0.82283 | 1.068422 | 0.835349 | -2.682934189 | -2.682934189 | -2.682934189 |
| 26 | RNA-binding protein 25 | 0.240599 | 0.642963 | 0.572124 | -3.238113765 | -3.238113765 | -3.238113765 |
| 27 | Splicing factor 3B subunit 2 | 0.270645 | 1.218281 | -0.54197 | -0.773399755 | -0.657034999 | -0.773399755 |
| 28 | Splicing factor U2af small subunit B | 0.454521 | 0.634509 | -0.44233 | -1.463101427 | -1.463101427 | -1.463101427 |
| 29 | Splicing factor U2AF 65 kDa subunit | 0.11815 | 0.929108 | 0.938241 | -0.780631873 | -0.79641487 | -0.79641487 |
| 30 | Protein RRC1 | 0.532613 | 0.082866 | -0.39497 | -1.366562985 | -1.366562985 | -1.366562985 |
| 31 | Pre-mRNA-splicing factor 8 homolog | 0.570287 | 0.259443 | -0.02415 | -3.086094827 | -3.086094827 | -3.086094827 |
| 32 | Pre-mRNA-processing-splicing factor 8A | -0.11281 | 0.637654 | 0.642344 | -2.907325618 | -2.907325618 | -2.907325618 |
| 33 | Pre-mRNA-splicing factor ISY1 homolog | 0.395342 | 0.524189 | -0.37189 | -0.371889402 | -0.371889402 | -0.371889402 |
| 34 | THO complex subunit 1 | 0.093394 | 0.446651 | -0.47707 | -0.477068528 | -0.477068528 | -0.477068528 |
| 35 | Serine/arginine-rich-splicing factor SR34 | -0.2751 | 0.380628 | 0.087468 | -1.088216087 | -1.088216087 | -1.088216087 |
| 36 | Serine/arginine-rich splicing factor SC35 | 0.733706 | 0.937683 | 0.670312 | -2.421557464 | -2.421557464 | -2.421557464 |
| 37 | Serine/arginine-rich splicing factor RS31 | 0.58339 | 0.093735 | 0.63455 | -0.452233488 | -0.452233488 | -0.452233488 |
| 38 | Serine/arginine-rich SC35-like splicing factor SCL33 | -0.1577 | 0.030388 | 0.045774 | -0.454007978 | -0.454007978 | -0.454007978 |
| 04012 ErbB signaling pathway (1) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 39 | Serine/threonine-protein kinase TOR | -0.13965 | 0.024261 | 0.222411 | -0.467335376 | -0.467335376 | -0.467335376 |
| 04016 MAPK signaling pathway – plant (8) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 40 | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 | -0.39322 | 0.158405 | 0.151183 | -0.981064495 | -0.981064495 | -0.981064495 |
| 41 | Transcription factor bHLH14 | 0.548965 | 0.884457 | -0.22924 | -0.229243769 | -0.229243769 | -0.229243769 |
| 42 | Transcription factor MYC2 | 0.463226 | 0.388223 | 0.246732 | -3.542892865 | -3.542892865 | -3.542892865 |
| 43 | Abscisic acid receptor PYL4 | 1.852722 | 2.016651 | 1.774202 | -2.549995805 | -3.405985503 | -2.273079691 |
| 44 | Abscisic acid receptor PYL9 | -0.04695 | 0.02713 | 0.14755 | -0.851006972 | -0.851006972 | -0.851006972 |
| 45 | Serine/threonine-protein kinase SAPK10 | 0.085518 | 0.408981 | 0.024526 | -0.672358315 | -0.672358315 | -0.672358315 |
| 46 | Serine/threonine-protein kinase SRK2I | 0.352227 | 0.265327 | 0.300548 | -0.464986643 | -0.464986643 | -0.464986643 |
| 47 | Ethylene-insensitive protein 2 | 0.625007 | 0.49946 | 0.634267 | -2.388455563 | -2.388455563 | -2.388455563 |
| 48 | ETHYLENE INSENSITIVE 3-like 3 protein | 0.006315 | 0.249024 | 0.482061 | -0.302861221 | -0.302861221 | -0.302861221 |
| 49 | Mitogen-activated protein kinase 3 | -0.32703 | -0.24247 | -0.65804 | -2.104820666 | -2.104820666 | -2.104820666 |
| 50 | Mitogen-activated protein kinase kinase kinase NPK1 | -0.10581 | 0.342507 | -0.21778 | -0.406812599 | -0.406812599 | -0.406812599 |
| 04072 Phospholipase D signaling pathway (3) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 51 | Dynamin-2B | 0.036532 | 0.427132 | -0.81227 | -0.812266541 | -0.812266541 | -0.812266541 |
| 52 | Serine/threonine-protein kinase TOR | -0.13965 | 0.024261 | 0.222411 | -0.467335376 | -0.467335376 | -0.467335376 |
| 53 | Ankyrin repeat- PH and SEC7 domain containing protein secG | 0.054366 | 0.039542 | 0.172577 | -0.679422307 | -0.679422307 | -0.679422307 |
| 04075 Plant hormone signal transduction (12) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 54 | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 | -0.39322 | 0.158405 | 0.151183 | -0.981064495 | -0.981064495 | -0.981064495 |
| 55 | Transcription factor bHLH14 | 0.548965 | 0.884457 | -0.22924 | -0.229243769 | -0.229243769 | -0.229243769 |
| 56 | Transcription factor MYC2 | 0.463226 | 0.388223 | 0.246732 | -3.542892865 | -3.542892865 | -3.542892865 |
| 57 | ABSCISIC ACID-INSENSITIVE 5-like protein 2 | 0.147921 | 0.100752 | 0.205535 | -0.336475436 | -0.336475436 | -0.336475436 |
| 58 | Ubiquitin C-terminal hydrolase 22 | 0.85731 | 0.59537 | -1.46606 | -1.466060125 | -1.466060125 | -1.466060125 |
| 59 | Auxin-responsive protein IAA14 | 1.958978 | 1.720019 | 1.690397 | -2.897808345 | -2.897808345 | -1.762930291 |
| 60 | Auxin-responsive protein IAA7 | -1.12006 | -1.0914 | -1.24864 | -2.252966449 | -2.250083941 | -2.251524475 |
| 61 | Protein TRANSPORT INHIBITOR RESPONSE 1 (TIR1) | 1.184285 | 1.267837 | 1.534964 | -2.01593676 | -2.01593676 | -2.01593676 |
| 62 | Auxin response factor 5 | -0.20576 | 0.035701 | -0.49736 | -0.497363523 | -0.497363523 | -0.497363523 |
| 63 | Auxin response factor 15 | -0.20652 | -0.18762 | -0.10993 | -2.070066629 | -2.070066629 | -2.070066629 |
| 64 | Two-component response regulator ARR3 | 0.66018 | 0.893755 | 0.665409 | -0.801348332 | -0.801348332 | -0.801348332 |
| 65 | Abscisic acid receptor PYL4 | 1.852722 | 2.016651 | 1.774202 | -2.549995805 | -3.405985503 | -2.273079691 |
| 66 | Abscisic acid receptor PYL9 | -0.04695 | 0.02713 | 0.14755 | -0.851006972 | -0.851006972 | -0.851006972 |
| 67 | Serine/threonine-protein kinase SAPK10 | 0.182149 | 0.394438 | 0.649098 | -0.879973008 | -0.879973008 | -0.879973008 |
| 68 | Serine/threonine-protein kinase SRK2I | 0.352227 | 0.265327 | 0.300548 | -0.464986643 | -0.464986643 | -0.464986643 |
| 69 | Methyltransferase-like protein 2 | 0.088466 | 0.244608 | 0.328877 | -0.652245369 | -0.652245369 | -0.652245369 |
| 70 | Ethylene-insensitive protein 2 | 0.625007 | 0.49946 | 0.634267 | -2.388455563 | -2.388455563 | -2.388455563 |
| 71 | ETHYLENE INSENSITIVE 3-like 3 protein | 0.006315 | 0.249024 | 0.482061 | -0.302861221 | -0.302861221 | -0.302861221 |
| 04136 Autophagy – other (3) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 72 | Serine/threonine-protein kinase TOR | -0.13965 | 0.024261 | 0.222411 | -0.467335376 | -0.467335376 | -0.467335376 |
| 73 | Autophagy-related protein 8C | 0.034356 | 0.272842 | -1.83552 | -1.835515633 | -1.831194027 | -1.58334222 |
| 04141 Protein processing in endoplasmic reticulum (9) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 74 | Protein transport protein Sec61 subunit beta | -0.41036 | -0.01659 | -1.17928 | -1.815265804 | -1.815265804 | -1.815265804 |
| 75 | Heat shock protein 90-5- chloroplastic | 1.960161 | 2.058363 | 1.856076 | -4.675500173 | -4.675500173 | -4.675500173 |
| 76 | Ubiquitin recognition factor in ER-associated degradation protein 1 | 0.284782 | 0.397391 | -0.11243 | -1.516790405 | -1.516790405 | -1.516790405 |
| 04150 mTOR signaling pathway (4) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 77 | CBL-interacting protein kinase 1 | 0.411691 | 0.2668 | 0.148233 | -1.62111525 | -1.62111525 | -1.62111525 |
| 78 | Serine/threonine-protein kinase TOR | -0.13965 | 0.024261 | 0.222411 | -0.467335376 | -0.467335376 | -0.467335376 |
| 79 | Serine/threonine-protein kinase ATG1c | -0.08368 | -0.07452 | 0.311552 | -0.581421762 | -0.581421762 | -0.581421762 |
| 80 | Calcium-binding protein 39-like | 0.015711 | 0.085252 | 0.051792 | -1.643756869 | -1.643756869 | -1.520752915 |
| 04151 PI3K-Akt signaling pathway (5) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 81 | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform | 0.810633 | 0.662534 | -0.48803 | -0.488025561 | -0.488025561 | -0.488025561 |
| 82 | CBL-interacting protein kinase 1 | 0.411691 | 0.2668 | 0.148233 | -1.62111525 | -1.62111525 | -1.62111525 |
| 83 | Serine/threonine-protein kinase TOR | -0.13965 | 0.024261 | 0.222411 | -0.467335376 | -0.467335376 | -0.467335376 |
| 84 | Myb-related protein A | 0.451647 | 0.311785 | 0.130007 | -2.191054894 | -2.191054894 | -2.191054894 |
| 85 | Heat shock protein 90-5- chloroplastic | 1.960161 | 2.058363 | 1.856076 | -4.675500173 | -4.675500173 | -4.675500173 |
| 04211 Longevity regulating pathway (2) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 86 | CBL-interacting protein kinase 1 | 0.411691 | 0.2668 | 0.148233 | -1.62111525 | -1.62111525 | -1.62111525 |
| 87 | Serine/threonine-protein kinase TOR | -0.13965 | 0.024261 | 0.222411 | -0.467335376 | -0.467335376 | -0.467335376 |
| 04213 Longevity regulating pathway – multiple species (2) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 88 | CBL-interacting protein kinase 1 | 0.411691 | 0.2668 | 0.148233 | -1.62111525 | -1.62111525 | -1.62111525 |
| 89 | Serine/threonine-protein kinase TOR | -0.13965 | 0.024261 | 0.222411 | -0.467335376 | -0.467335376 | -0.467335376 |
| 04218 Cellular senescence (3) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 90 | Calcineurin B-like protein 3 | 1.070011 | 1.306436 | 1.147796 | -1.734456047 | -1.734456047 | -1.734456047 |
| 91 | Serine/threonine-protein kinase TOR | -0.13965 | 0.024261 | 0.222411 | -0.467335376 | -0.467335376 | -0.467335376 |
| 92 | Mitochondrial outer membrane protein porin 2 | 0.659578 | 0.668525 | 0.995054 | -0.288275387 | -0.288275387 | -0.288275387 |
| 04370 VEGF signaling pathway (1) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 93 | Calcineurin B-like protein 3 | 1.070011 | 1.306436 | 1.147796 | -1.734456047 | -1.734456047 | -1.734456047 |
| 04530 Tight junction (3) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 94 | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform | 0.810633 | 0.662534 | -0.48803 | -0.488025561 | -0.488025561 | -0.488025561 |
| 95 | CBL-interacting protein kinase 1 | 0.411691 | 0.2668 | 0.148233 | -1.62111525 | -1.62111525 | -1.62111525 |
| 96 | Tubulin alpha-5 chain | 0.591109 | -0.00636 | 0.338417 | -2.790866288 | -2.790866288 | -2.790866288 |
| 04630 JAK-STAT signaling pathway (1) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 97 | Serine/threonine-protein kinase TOR | -0.13965 | 0.024261 | 0.222411 | -0.467335376 | -0.467335376 | -0.467335376 |
| 04710 Circadian rhythm (1) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 98 | CBL-interacting protein kinase 1 | 0.411691 | 0.2668 | 0.148233 | -1.62111525 | -1.62111525 | -1.62111525 |
| 04714 Thermogenesis (9) | N1 | N2 | N3 | NR1 | NR2 | NR3 | |
| 99 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit 1- mitochondrial | 1.353688 | 0.91732 | 1.373706 | -1.736656898 | -1.736656898 | -1.736656898 |
| 100 | Cytochrome c oxidase assembly protein COX11- mitochondrial | 1.118139 | 0.44765 | 0.259696 | -0.848660867 | -0.848660867 | -0.848660867 |
| 101 | Cytochrome c oxidase assembly protein COX15 | 1.006468 | 0.719522 | 0.640444 | -0.994613871 | -0.994613871 | -0.994613871 |
| 102 | NADH dehydrogenase [ubiquinone] flavoprotein 1- mitochondrial | -0.18283 | 0.628526 | -0.4885 | -0.488504281 | -0.488504281 | -0.488504281 |
| 103 | CBL-interacting protein kinase 1 | 0.411691 | 0.2668 | 0.148233 | -1.62111525 | -1.62111525 | -1.62111525 |
| 104 | Serine/threonine-protein kinase TOR | -0.13965 | 0.024261 | 0.222411 | -0.467335376 | -0.467335376 | -0.467335376 |
| 105 | SWI/SNF complex subunit SWI3C | 0.082451 | 0.391757 | -0.14529 | -0.145290342 | -0.145290342 | -0.145290342 |
| 106 | PsbP domain-containing protein 7- chloroplastic | 0.099498 | 0.212171 | 0.147611 | -0.243057157 | -0.243057157 | -0.243057157 |
| 107 | Protein arginine methyltransferase NDUFAF7 homolog- mitochondrial {ECO:0000250|UniProtKB:Q7L592} | 0.684605 | 0.685377 | 0.493703 | -2.538045118 | -2.006975625 | -2.538045118 |
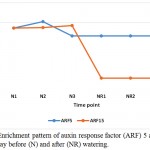 |
Figure 2: Enrichment pattern of auxin response factor (ARF) 5 and 15 at midday before (N) and after (NR) watering.
|
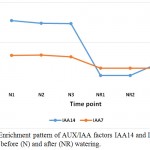 |
Figure 3: Enrichment pattern of AUX/IAA factors IAA14 and IAA7 at midday before (N) and after (NR) watering.
|
Aux/IAA proteins orchestrate several biological and physiological processes such as embryogenesis, leaf expansion and senescence, lateral root development and fruit development by regulating the expression of auxin response genes (Wilmoth et al. 2005, Sagar et al. 2013). IAA7 controls the morphological responses induced by light, e.g., promoting leaf development under adverse condition, while IAA14 acts in controlling lateral root formation by interacting with two ARF proteins (Luo et al. 2018). These two Aux/IAAs are produced due to the accumulated IAA generated is a result of tryptophan metabolism pathway and specifically oriented towards the induction of plant hormone signal transduction pathway to help the plant maintains normal performance inder drought stress. As the highly conserved domain II of the other Aux/IAA proteins is a target for degradation process promoted by auxin, this action allows the participation of the auxin-induced Protein TRANSPORT INHIBITOR RESPONSE 1 (TIR1). Consequently, TIR1 activates its target factors, e.g., ARFs, thus, allowing the auxin-responsive downstream genes, AUX/LAX, LBD and SAUR, to function and promote plant growth under adverse conditions. This Protein TRANSPORT INHIBITOR RESPONSE 1 (TIR1) also shown to be a major player under drought stress in senna as it is enriched only under the stress (Table 3, Figure 4). BRASSINOSTEROID INSENSITIVE 1 (BRI1)-associated receptor kinase 1 (or BAK1) belongs to a large group of plant transmembrane proteins known as the leucine-rich repeat receptor kinases (Belkhadir and Jaillais 2015). BAK1 has important role in brassinosteroid signaling (Li et al. 2002). Brassinosteroid is a plant hormone with important roles in cell proliferation and growth (Belkhadir and Jaillais 2015). BAK1 particularly acts in repressing the development of unneeded stomata in plant leaves, thus maintain water turgor under drought stress (Smakowska-Luzan et al. 2018). BAK1 also shown to be a major player under drought stress in senna as it is enriched only under the stress (Table 3, Figure 5).
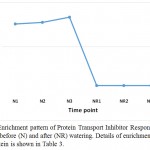 |
Figure 4: Enrichment pattern of Protein Transport Inhibitor Response 1 (TIR1) at midday before (N) and after (NR) watering. Details of enrichment pattern of this protein is shown in Table 3.
|
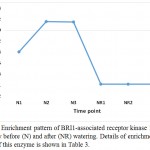 |
Figure 5: Enrichment pattern of BRI1-associated receptor kinase 1 (BAK1) at midday before (N) and after (NR) watering. Details of enrichment pattern of this enzyme is shown in Table 3.
|
In senna, it is evident that PYL and BAK1 are induced by ABA and brassinosteroid towards the occurrence of stomatal closure and in cell elongation and division. These roles represent two bottle-necks in the plant hormone signal transduction pathway. Over and above, indole-3-pyruvate (or flavin) monooxygenase, encoded by YUC2 gene, also participates in tryptophan metabolism pathway (Watanabe and Lam 2006) towards the biosynthesis of IAA that, as mentioned above, is required for triggering the plant hormone signal transduction pathway. Accordingly, this enzyme likely has a role in conferring drought stress tolerance in senna (C. angustifolia) (Table 3, Figure 6). The two pathways likely crosstalk in senna (C. angustifolia) towards the maintenance of normal growth under adverse condition (Figures 7 and 8).
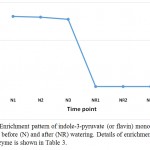 |
Figure 6: Enrichment pattern of indole-3-pyruvate (or flavin) monooxygenase at midday before (N) and after (NR) watering. Details of enrichment pattern of this enzyme is shown in Table 3.
|
 |
Figure 7: Tryptophan metabolism pathway indicating two important enzymes Aldehyde dehydrogenase family 3 member F1 (EC: 1.2.1.3) and indole-3-pyruvate monooxygenase (EC: 11.41.31.68) that were enriched under drought stress, while suppressed due to watering. Details of enrichment pattern of these two enzymes are shown in Table 3.
|
 |
Figure 8: Plant hormone signal transduction pathway indicating the auxin response factor (ARF) and the Aux/IAA that were enriched under drought stress, while suppressed due to watering. Details of enrichment pattern of these two enzymes are shown in Table 3.
|
Phosphoinositide phosphatase SAC1 and SAC6 can also be among the molecular mechanisms to cope with the stress in senna (C. angustifolia) (Table 3, Figure 9). The domains of SAC protein possess phosphoinositide phosphatase activities. SAC1 was previously shown to affect diverse cellular functions such as normal cell morphogenesis, Golgi function, and maintenance of vacuole morphology (Zhong et al. 2005), while SAC6 gene was predominantly expressed in flowers and proven to be highly induced by salinity in Arabidopsis (Zhong and Ye 2003).
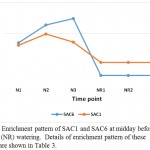 |
Figure 9: Enrichment pattern of SAC1 and SAC6 at midday before (N) and after (NR) watering. Details of enrichment pattern of these proteins are shown in Table 3.
|
A number of five pre-mRNA splicing factors as well as three serine/arginine-rich (SR) regulator factors were shown to participate in drought stress tolerance in senna (C. angustifolia) (Table 3, Figure 10). The pre-mRNA factors included splicing factors (SF) 8, 8A, 19, 40A and ISY1, while SRs included factors SCL33, RS31 and RS34. No particular function is assigned to any of these splicing factors and regulators except that each one might act on a certain protein or protein type. This conclusion warrant further experimentation and analysis. Pre-mRNAs usually contain introns that requires splicing or alternative splicing (AS) to produce structurally and functionally different proteins from the same gene (Palusa et al. 2007). Pre-mRNAs-related genes encode serine/arginine-rich (SR), which is a conserved protein family of splicing regulators in plant. Splicing of SR genes has proven to be developmental- and tissue-specific. Prior research indicated that abiotic stresses regulate the splicing of the pre-mRNAs of SR genes to produce different protein isoforms, thus different functions. We speculate that the altered splicing in senna seems like the action of the immune systems as to produce a specific antibodies for a specific antigen. Here, we think that alternative splicing of SR under stress produce the proper isoform of the proteins that can hold their structures and avoid denaturation or unfolding as a response to stimuli.
 |
Figure 10: Enrichment pattern of the five Pre-mRNA splicing factors SF19, SF A40, SF 8, SF 8A and SF ISY1 as well as the three serine/arginine-rich SR34, SC35 and SR31 regulator factors at midday before (N) and after (NR) watering. Details of enrichment pattern of these proteins are shown in Table 3.
|
Many other regulated metabolites and proteins in senna (C. angustifolia) including protective proteins, such as heat shock protein 95s were previously reported to play functional roles in drought tolerance in plants (Hu and Xiong 2014, Umezawa et al. 2006). Senna might also developed efficient signal transduction cascades to cope with drought stress as it proved to regulate ATPase, which is a major signaling factor involved in drought stress signaling (Akpinar et al. 2012, 2013). Protein kinases participate in developmental and environmental signal transduction in plants (Rodriguez et al. 2010, Liu et al. 2016) and play a key role in activating transcription factors and drought-responsive proteins under drought tolerance. Among them, mitogen-activated protein kinase (MAPK) and cytochrome c oxidase that were regulated in senna (C. angustifolia) (Table 3).
Other enzymes include phospholipase C2 and allene oxide cyclase as well as isochorismate pathway were regulated in senna (C. angustifolia). The two enzymes play a role in JA pathway, while isochorismate pathway results in the production of salicylic acid (SA) (Kawano et al. 2004, Mustafa et al. 2009). The latter pathways have important applications in plant production.
In conclusion, we speculate that we have expanded our understanding of the molecular mechanism underlying drought stress tolerance in senna (C. angustifolia). This information will be valuable resource for accelerated genomics-assisted genetic breeding programs targeting the improvement of drought tolerance in economically important crop plants.
References
- Akpinar BA, Avsar B, Lucas SJ, Budak H (2012) Plant abiotic stress signalling. Plant Signal Behav 7(11):1450–1455.
CrossRef - Akpinar BA, Lucas SJ, Budak H (2013) Genomics approaches for crop improvement against abiotic stress. Sci World J 15:361921.
CrossRef - Ayoub AT (1977) Some primary features of salt tolerance in senna (Cassia angastifolia). J Exp Bot 28:484–492.
CrossRef - Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Cr Rev Plant Sci 24(1):23–58.
CrossRef - Belkhadir Y, Jaillais Y (2015). The molecular circuitry of brassinosteroid signaling”. The New Phytologist 206): 522–40.
CrossRef - Blum A (1988) Plant breeding for stress environments. CRC Press, Inc., Boca Raton.
- Bolger AM, LohseM, Usadel B (2014) Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120.
CrossRef - Bouzroud S, Gouiaa S, Hu N, Bernadac A, Mila I, Bendaou N, et al. (2018). Auxin Response Factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS ONE 13(2): e0193517.
CrossRef - Bowman MJ, Park W, Bauer PJ, Udall JA, Page JT, Raney J, Scheffler BE, Jones DC, Campbell BT (2013) RNA-Seq transcriptome profiling of upland cotton (Gossypium hirsutum L.) root tissue under water-deficit stress. PLoS One 8(12):e82634.
CrossRef - Budak H, Kantar M, Kurtoglu KY (2013) Drought tolerance in modern and wild wheat. Sci World J 15:548246.
CrossRef - DeJong M, Wolters-Arts M, Schimmel BC, Stultiens CL., de Groot PF, et al. (2015). Solanum lycopersicum AUXIN RESPONSE FACTOR 9 regulates cell division activity during early tomato fruit development. J. Exp. Bot.66, 3405–3416.
CrossRef - Do PT, Degenkolbe T, Erban A, Heyer AG, Kopka J, Köhl KI, Hincha DK, Zuther E (2013) Dissecting rice polyamine metabolism under controlled long-term drought stress. PLoS One 8(4):e60325.
CrossRef - Dong Y, Fan G, Deng M, Xu E, Zhao Z (2014) Genome-wide expression profiling of the transcriptomes of four Paulownia tomentosa accessions in response to drought. Genomics 104(4):295–305.
CrossRef - Ergen NZ, Thimmapuram, Bohnert J, Hans J, Budak H (2009) Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct Integr Genomics 9(3):377–396.
CrossRef - Folkard C (1995) Encyclopedia of herbs and their uses. Herb Society of America, Dorling Kindersley Publishing Inc., New York.
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414: 271-276.
CrossRef - Hammouda FM, Ismail SI, Abdel-Azim NS, Shams KA (2005) A guide to medicinal plants in North Africa. In: Batanouny KH (eds) IUCN Centre for Mediterranean Cooperation, Malaga, Andalusia, Spain, pp 217–218.
- Hao Q., Yin P., Li W., Wang L., Yan C., et al. (2011). The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol. Cell 42: 662-672.
CrossRef - Hirayama T, Shinozaki K (2010) Research on plant abiotic stress responses in the postgenome era: past, present and future. Plant J 61(6):1041–1052.
CrossRef - Hiz MC, Canher B, Niron H, Turet M (2014) Transcriptome analysis of salt tolerant common bean (Phaseolus vulgaris L.) under saline conditions. PLoS One 9(3):e92598.
CrossRef - Hu H, Xiong L (2014) Genetic engineering and breeding of droughtresistant crops. Annu Rev Plant Biol 65:715–741.
CrossRef - Hu T, Sun X, Zhang X, Nevo E, Fu J (2014) An RNA sequencing transcriptome analysis of the high-temperature stressed tall fescue reveals novel insights into plant thermotolerance. BMC Genomics 15:1147.
CrossRef - Kang C, He S, Zhai H, Li R, Zhao N and Liu Q (2018) A Sweetpotato Auxin Response Factor Gene (IbARF5) Is Involved in Carotenoid Biosynthesis and Salt and Drought Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 9: 1307.
CrossRef - Kantar M, Lucas SJ, Budak H (2011) Drought stress: molecular genetics and genomics approaches. Adv Bot Res 57:445–493.
CrossRef - Kawano T, Furuichi T, Muso S (2004) Controlled free salicylic acid levels and corresponding signaling mechanisms in plants. Plant Biotechnol 21:319–335.
CrossRef - Khalid H, Abdalla WE, Abdelgadir H, Opatz T, Effert T (2012) Gems from traditional north-African medicine: medicinal and aromatic plants from Sudan. Nat Prod Bioprospect 2(3):92–103.
CrossRef - Khammari I, Galavi M, Ghanbari A, Solouki M, Poorchaman MRA (2012) The effect of drought stress and nitrogen levels on antioxidant enzymes, proline and yield of Indian Senna (Cassia angustifolia L.). J Med Plants Res 6(11):2125–2130.
CrossRef - Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI (2006). The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140(1):127-39.
CrossRef - Lee SC, Lan W, Buchanan BB, Luan S (2009). A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA. 106(50):21419-24.
CrossRef - Lemli J (1986) The chemistry of senna. Fitoterapia 57:33–40.
- Levitt J (1980) Responses of plants to environmental stress: chilling, freezing and high temperature stresses, vol 1, 2nd edn. Academic, New York.
CrossRef - Li J, Wen J, Lease KA, Doke JT, Tax FE, et al. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–22.
CrossRef - Li MY, Tan HW, Wang F, Jiang Q, Xu ZS, et al. (2014a) De novo transcriptome sequence assembly and identification of AP2/ERF transcription factor related to abiotic stress in parsley (Petroselinum crispum). PLoS One 9(9):e108977.
CrossRef - Li PS, Yu TF, He GH, Chen M, Zhou YB, et al. (2014b) Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genom 15:1009.
CrossRef - Li WX, Oono Y, Zhu J, He XJ, Wu JM, et al. (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and post-transcriptionally to promote drought resistance. Plant Cell 20(8):2238–2251.
CrossRef - Liu H, Che Z, Zeng X, Zhou X, Sitoe HM, et al. (2016) Genome-wide analysis of calcium-dependent protein kinases and their expression patterns in response to herbivore and wounding stresses in soybean. Funct Integr Genom 16(5):481–493.
CrossRef - Luo J, Jing-Jing Zhou J-J, Zhang J-Z (2018). Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int J Mol Sci. 19(1): 259.
CrossRef - Mehta RH, Ponnuchamy M, Kumar J, et al. (2017). Exploring drought stress-regulated genes in senna (Cassia angustifolia Vahl.): a transcriptomic approach. Funct Integr Genomics (2017) 17:1–25.
CrossRef - Mustafa NR, Kim HK, Choi YH, Erkelens C, Lefeber AW, Spijksma G, van der Heijden R,Verpoorte R (2009) Biosynthesis of salicylic acid in fungus elicited Catharanthus roseus cells. Phytochemistry 70(4): 532–539.
CrossRef - Palusa SG, Ali GS, Reddy AS (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49(6):1091-107.
CrossRef - Pizzio GA, Rodriguez L, Antoni R, Gonzalez-Guzman M, Yunta C, et al. (2013). The PYL4 A194T Mutant Uncovers a Key Role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA Interaction for Abscisic Acid Signaling and Plant Drought Resistance. Plant Physiol. 163(1): 441–455.
CrossRef - Rama Reddy NR, Ragimasalawada M, Sabbavarapu MM, Nadoor S, Patil JV (2014) Detection and validation of stay-green QTL in post-rainy sorghum involving widely adapted cultivar, M35-1 and a popular stay-green genotype B35. BMC Genom 18(15):909.
CrossRef - Ramchandra Reddy A, Chaitanya KV, Vivekanandan M (2004) Drought induced responses of photosynthesis and antioxidant metabolism in higher plants. J Plant Physiol 161(11):1189–1202.
CrossRef - Remington DL, Vision TJ, Guilfoyle TJ, Reed JW (2004). Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135: 1738-1752
CrossRef - Ratnayaka HH, Kincaid D (2005) Gas exchange and leaf ultrastructure of tinnevelly senna, cassia angustifolia, under drought and nitrogen stress. Crop Sci 45(3):840–847.
CrossRef - Rocheta M, Becker JD, Coito JL, Carvalho L, Amâncio S (2014) Heat and water stress induce unique transcriptional signatures of heatshock proteins and transcription factors in grapevine. Funct Integr Genomics 14(1):135–148.
CrossRef - Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61:621–649.
CrossRef - Sagar M, Chervin C, Mila I, Hao Y, Roustan JP, et al. (2013). SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 161(3):1362-74.
CrossRef - Shanker AK, Maheswari M, Yadav SK, Desai S, Bhanu D, Attal NB, Venkateswarlu B (2014) Drought stress responses in crops. Funct Integr Genomics 14(1):11–22.
CrossRef - Smakowska-Luzan E, Mott GA, Parys K, Stegmann M, Howton TC, et al. (2018). An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553: 342–346.
CrossRef - Thumma BR, Sharma N, Southerton SG (2012) Transcriptome sequencing of Eucalyptus camaldulensis seedlings subjected to water stress reveals functional single nucleotide polymorphisms and genes under selection. BMC Genomics 13:364.
CrossRef - Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr Opin Biotechnol 17(2): 113–122.
CrossRef - Watanabe N, Lam E (2006). Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. The Plant Journal. 45, 884-894.
CrossRef - Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, et al. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43(1):118-30.
CrossRef - Xu Y, Gao S, Yang Y, Huang M, Cheng L,Wei Q, Fei Z, Gao J, Hong B (2013) Transcriptome sequencing and whole genome expression profiling of chrysanthemum under dehydration stress. BMC Genomics 14:662.
CrossRef - Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular response and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803.
CrossRef - Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, et al. (2006). ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 140(1):115-26.
CrossRef - Zhang J, Ruhlman TA, Mower JP, Jansen RK (2013b) Comparative analyses of two Geraniaceae transcriptomes using next-generation sequencing. BMC Plant Biology 13: 228.
CrossRef - Zhang X, Allan AC, Li C,Wang Y, Yao Q (2015) De novo assembly and characterization of the transcriptome of the Chinese medicinal herb, Gentiana rigescens. Int J Mol Sci 16(5):11550–11573.
CrossRef - Zhao Y, Chan Z, Gao J, Xing L, Cao M, et al. (2016). ABA receptor PYL9 promotes drought resistance and leaf senescence. PNAS 113(7):1949-1954.
CrossRef - Zhong R, Burk DH, Nairn CJ, Wood-Jones A, Morrison WH, et al. (2005). Mutation of SAC1, an Arabidopsis SAC domain phosphoinositide phosphatase, causes alterations in cell morphogenesis, cell wall synthesis, and actin organization. Plant Cell 17:1449-1466.
CrossRef - Zhong R, Ye Z-H (2003). The SAC Domain-Containing Protein Gene Family in Arabidopsis. Plant Physiol. 132(2): 544–555.
CrossRef - Zhou X, Li L, Xiang J, Gao G, Xu F, Liu A, Zhang X, Peng Y, Chen X, Wan X (2015) OsGL1-3 is involved in cuticular wax biosynthesis and tolerance to water deficit in rice. PLoS One 10(1):e116676.
CrossRef - Zhu H, Dardick CD, Beers EP, Callanhan AM, Xia R, Yuan R (2011) Transcriptomics of shading-induced and NAA-induced abscission in apple (Malus domestica) reveals a shared pathway involving reduced photosynthesis, alterations in carbohydrate transport and signaling and hormone crosstalk. BMC Plant Biol 11:138.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





