Manuscript accepted on : 27/12/2019
Published online on: --
Plagiarism Check: Yes
Reviewed by: Jean-Paul MOTTA
Second Review by: kavitha kandimalla
Subcloning and Expression of a Protease Gene from Bacillus Halodurans CM1 in Bacillus Subtilis DB104
Endang Rahmawati1, Abinawanto1 and Is Helianti2*
1Department of Biology, Faculty of Mathematics and Natural Science, University of Indonesia, Depok, Jawa Barat, Indonesia.
2Center for Bioindustrial Technology, Agency of Assessment and Application of Technology (BPPT), 611/614 Building, LAPTIAB-BPPT, PUSPITEK-Serpong, Tangerang Selatan 15314, Indonesia.
Corresponding Author E-mail: is.helianti@bppt.go.id
DOI : http://dx.doi.org/10.13005/bbra/2799
ABSTRACT: Proteases are potential enzymes that utilized in various industrial fields, and the demand of these enzymes is increasing. Bacillus halodurans CM1 is Indonesia indigenous bacterium which is detected to be able to produce alkalotermophilic protease enzyme. In this study, we subcloned the protease gene consist of Open Reading Frame of protease gene and its promoter from Bacillus halodurans CM1 in Bacillus subtilis DB104 via conjugation, and analyzed the expression of the recombinant protease. The protease gene is 1 417 bp length including the open reading frame and the promoter, and obtained by PCR and cloned into pGEM T easy. After confirmed by sequencing, the gene was subcloned into vector pBBRE194, then the recombinant plasmid was transformed into E. coli S17-1. This E.coli was then conjugated to Bacillus subtilis DB104. The target recombinant B. subtilis DB104 has been obtained confirmed by plasmid verification and erythromycin resistance. The recombinant protease produced showed the highest enzyme activity at 50oC and pH 9 (with pH range 5-9) which with protease activity 13.66 U/mL.
KEYWORDS: Bacillus Halodurans CM1; Bacillus Subtlis DB104; Conjugation; Expression; Protease Gene; Subcloning
Download this article as:| Copy the following to cite this article: Rahmawati E, Abinawanto, Helianti I. Subcloning and Expression of a Protease Gene from Bacillus Halodurans CM1 in Bacillus Subtilis DB104 .Biosci Biotech Res Asia 2019;16(4). |
| Copy the following to cite this URL: Rahmawati E, Abinawanto, Helianti I. Subcloning and Expression of a Protease Gene from Bacillus Halodurans CM1 in Bacillus Subtilis DB104 .Biosci Biotech Res Asia 2019;16(4). Available from: https://bit.ly/2T34GbH |
Introduction
Proteases are proteolytic enzyme that catalyze the breakdown of proteins through hydrolysis of peptide bonds. Proteases enzyme are ubiquitous and being found all living organism because is crucial for cell growth and differentiation [1, 2]. They can be produced by various type of organisms, including bacteria, actinomycetes, yeast, mold, and animal tissues [3]. The use of microorganisms such as bacteria to produce enzyme has many advantages. These advantages are easily produced on a large scale, relatively short production time, and can be produced sustainably at low cost.[4]
Proteases have high commercial value in the industrial fields. They widely used in the food, animal feed, pharmaceutical, leather tanning, and detergent industries. Moreover, proteases are one of the largest selling enzymes in the world, accounting for 60% of the total commercialized enzymes [5, 6]. Industries generally use protease produced by bacteria. The type of bacteria that is widely used for protease production comes from the genus Bacillus [7]. Several studies of protease gene cloning have been reported from Bacillus pumilus, Bacillus halodurans JB99, and Bacillus licheniformis 2709 [8, 9, 10]. Several species of Bacillus sp. are also known to have the ability for producing alkalothermophilic protease. This protease active at pH>7 and resistant in high temperatures [11], so that make it suitable for harsh industrial processes. Therefore, many studies have been conducted to find new isolates as producer of alkalotermophilic protease enzymes.
Bacillus halodurans CM1 is Indonesia indigenous alkalotermophilic bacteria, that was successfully isolated by the Center for Bioindustrial Technology, BPPT from hot spring sediment, Cimanggu, West Java. Based on the analysis of 16S rRNA gene and biochemical characteristics, it was identified as a new strain with 99% similarity to B. halodurans C-125 [12]. Bacillus halodurans C-125 is one of the strains of species B. halodurans whose whole genome has been readily determined [13, 14]. Bacillus halodurans CM1 was identified as a new strain, because it has different morphological and biochemical characters with B. halodurans C-125.
Bacillus halodurans CM1 produces potential xylanase enzyme and show activity of other enzymes, such as protease, amylase, and gelatinase [12]. Cloning of the ORF of protease gene from B. halodurans in E. coli DH5α has been reported by Helianti [15]. However, the cloning of the whole gene with the promoter and expressed in Bacillus subtilis has not been reported yet. This protease gene consists of the open reading frame (ORF) and the original promoter region (upstream) measuring 1 411 bp. It had 99% similarity with alkaline thermostable protease of B. halodurans.
Bacillus halodurans CM1 has great potential, because it has not been genetically engineered (wild type) and commercialized. It can also produce alkalotermophilic protease that make it promising opportunity to be developed and utilized in the industrial scale. Therefore, in this study we subcloned the protease gene from B. halodurans CM1, then transformed it to Bacillus subtilis DB104 via conjugation and analyzed the expression of the recombinant gene product.
Material and Methods
Strains, Plasmids, and Media
The bacterial strain originally used as host for obtaining and maintaining the recombinant plasmid was E. coli DH5α. Escherichia coli S17-1 served as the donor and Bacillus subtilis DB104 as the recipient cell in conjugation experiments. The plasmid DNA of pGEM-T easy containing protease gene from B. halodurans CM1 served as the source of the protease gene and the plasmid DNA of pBBRE cell-A served as the vector. Cloning vector pBBRE194 used is conjugative shuttle vector of E. coli–Bacillus. It was constructed from pE194 and PBBR MCS3, that carries two origins of replication as well as two antibiotic resistant genes (erythromycin and tetracycline) [16]. All strains and plasmids are collection of Center for Bioindustrial Technology, Agency of Assessment and Application of Technology (BPPT). LB and LB-skim milk media with tetracycline (12.5 µg/mL) was used to select E. coli DH5α transformants, or erythromycin (5 µg/mL) was used to select B. subtilis DB104 transconjugants.
DNA Extraction and Primers Design
Plasmid DNA from the recombinant E. coli DH5α harboring plasmid pGEM-T easy prot-CM1 and the recombinant E. coli DH5α harboring plasmid pBBRE194 cell-A were extracted by using small scale alkali lyses method. Plasmid pGEM-T easy prot-CM1 was used as PCR template of the protease gene and plasmid pBBRE194 cell-A was used as the source of plasmid pBBRE194 by cutting it with KpnI and PstI. All genetic experiments were performed according to the protocols in Sambrook and Russel [17]. The protease gene was amplified by using a pair of oligonucleotides, 5’-AAG GGT ACC GAA ATC GAG CTA TAG GGA GCG AAC-3’ as the forward and 5’-GTT CTG CAG CTA TTG TGT TGC ACC TGC AGC ATG-3’ as the reverse primer. Concomitantly designed KpnI and PstI restriction sites, which were used for cloning, are underlined. Primers were designed manually by analyzing sequence of the protease gene from B. halodurans CM1 with strictly considering to the temperature melting and GC content.
Polymerase Chain Reaction (PCR)
After the initial denaturation 30 s at 98 oC, the mixture was subjected to 30 cycles, each consisting of 10 s denaturation at 98 °C, 15 s annealing at 65 °C, and 45 min extension at 72 °C, followed by 10 min final extention at 72 °C to complete the elongation. The thermal cycler from Eppendorf (Germany) and the High Fidelity Phusion DNA Polymerase (Thermo scientific, UK) were used. The amplified fragment was purified using the Geneaid PCR Clean Up Kit (Geneaid, China), then cut it with KpnI and PstI and subsequently ligated into pBBRE194, which was linearized by the same restriction enzymes. The ligation mixture was used to transform E. coli DH5α. The procedure of cloning and transformation is shown schematically in Fig 1.
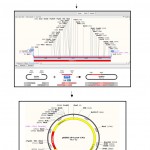 |
Figure 1: Schematic representation of the cloning procedure of protease gene from Bacillus halodurans CM1 and conjugation to Bacillus subtilis as the recipient cell. |
Bacterial Conjugation
The recombinant plasmid pBBRE194 prot-CM1 was isolated from recombinant E. coli DH5α and retransformed into E. coli S17-1, which would serve as the donor for the conjugative transfer. The conjugation process was carried out to B. subtilis DB104 as the recipient cell. The bacterial conjugation from the recombinant E. coli S17-1 to B. subtilis DB104 was conducted based on the protocol developed by Aquino de Muro and Priest [18], which was optimized by Richhardt et al. [19] with considering to Helianti [16]. For conjugation process, B. subtilis DB104 cells were initially grown in LB broth without antibiotics, whereas E. coli S17-1 harboring the recombinant plasmid pBBRE194 prot-CM1 was cultivated at 30ºC over night in LB media with tetracycline. Two 200-ml Erlenmeyer flasks, each containing 27 ml of sporulation medium, was subsequently inoculated with either 3 ml of the overnight B. halodurans CM1 or E. coli culture. The cultures were then grown at 30ºC until OD600 nm reached 0.6-0.8. The cells were then harvested by centrifugation (3000 x g, 4ºC) and washed twice in 15 ml holding buffer (12. 5 mM KH2PO, 12.5 mM K2HPO4, 1 mM MgSO4, pH 7.2) and pelleted by centrifugation. After resuspension in the holding buffer, the recipient cells are mixed with the donor cells. Using syringe and filter, the mixture was compressed on a sterile nitrocellulose 0.45 µm filter to ensure close contact of donor and recipient cells. The filter was placed on sporulation agar for 48 h at 30ºC with the side containing cells facing upwards [20]. Counter selection of B. subtilis DB104 transconjugants against E. coli was performed by pasteurization, where cells were collected from the filter by suspending in 1 mL holding buffer and then incubated for 20 min at 80 ºC, before subsequently spread on LB agar plates containing erythromycin. The transconjugants were further analyzed with respect to their protease activity and gene verification by restriction and PCR analysis.
Enzyme Preparations from Recombinant B. subtilis DB104
A single recombinant B. subtilis DB104 colony harboring pBBRE194 prot-CM1 was used to inoculate in 25 mL media (LB containing erythromycin) and grown overnight. The culture (3 mL) was then transferred into an Erlenmeyer flask with 30 ml media and shaken at 150 rpm, 37 °C in a Kühner Shaker (Kühner, Switzerland). The culture was grown until OD600nm reached 0.6-0.8. As control, a single B. subtilis DB104 colony without plasmid was used as inoculum and subjected to the same procedure, with media without antibiotic. All cultures must have same optical density to be starter for enzyme production.
The recombinant Bacillus subtilis DB104 starter was added in the production medium, which is 50 mL medium (LB and LB with 4% corncob) containing erythromycin 5 mg/mL. Meanwhile, 5 mL of Bacillus subtilis DB104 starter was also add in 50 mL media (LB and LB with 4% corncob) without antibiotics. The production culture is homogenized with the starter, then each production culture is sampled 1 mL to measure the initial OD of the production culture before fermentation. Furthermore, the production culture was fermented in a shaker incubator at a speed of 150 rpm at 37 oC for 24 hours. For harvesting, the production culture was centrifugated at 4 oC at rate of 8 000 x g for 30 minutes, so was obtained crude extract enzyme.
Enzymatic Activity Assay
The protease activity was measured (each sample in triplicate) by the method from Amano 2007 [21], using folin reagent and sodium carbonate to quantify the caseins reduced. Tyrosine was used as standard. A unit of protease activity is the number of enzymes that liberate amino acids and peptides. In this case, the value corresponds to the number of enzymes liberating 1μg of tyrosine per minute (from casein) under test conditions (U/mL). Comparison of protease enzyme activities between the recombinant B. subtilis DB104 and B. subtilis DB104 was measured at temperature of 50 °C and pH 9. The protease enzyme activity was measured using a spectrophotometer at a wavelength of 660 nm. Characterization of temperature on the protease activity was measured (each sample in triplicate) at temperature range between 30-60 °C, pH 9. Characterization of pH on the protease activity was measured (each sample in triplicates) in a range of 5-9 at 50 °C [22]. For measuring enzyme activity, was used 50 mM of the following buffers; Na-citrate buffer (for pH 5-6), Na-phosphate buffer (for pH 7), and Tris-HCl buffer (pH 8-9).
Protein Assay
Protein content was determined using the method of Bradford 1976 [23]. Bovine Serum Albumin (BSA) Fraction V was used as a protein for preparing the standard solutions. The samples were tested by mixing 20 mL of enzyme and 1 mL of Bradford reagent, then being examined and allowed to stand for 15 minutes at room temperature. Meanwhile, the blank was tested by mixing 20 µL milli-Q sterile and 1 mL Bradford reagent, then left for 15 minutes at room temperature. After 15 minutes, the sample and blank were measured by the spectrophotometer at a wavelength of 595 nm. Readable absorbance is compared to the standard protein curve obtained.
Results and Discussion
We successfully amplified the specific 1417 bp DNA fragment using primers designed based on the protease gene from B. halodurans CM1. The protease gene consisted of the open reading frame (ORF) and the original promoter region. This specific fragment was then cloned in pBBRE194 vector of 8402 bp, and produced a recombinant DNA with 9 819 bp length namely plasmid pBBRE194 prot-CM1.
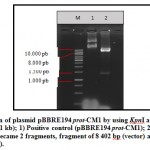
Plasmid pBBRE194 prot-CM1 was transformed to E. coli DH5α using the heat shock method [17]. This transformation resulted two clones grew in the selection media, which is LB media containing tetracycline. The transformants who grew well in LB containing tetracycline indicated that the plasmid pBBRE194 prot-CM1 has successfully entered in the cell. Plasmid pBBRE194 prot-CM1 is a plasmid carrying the tetr gene from E. coli, so if it is in the host cell of E. coli will be able to live and be resistant in the media containing tetracycline. Two clones of the transformants were then verified through plasmid extraction and restriction (Fig 2). The results of the plasmid restriction showed that the plasmid was successfully cut into 2 specific DNA fragments by KpnI and PstI with 8,402 and 1,417 bp length. DNA fragment of 8.402 bp is plasmid pBBRE194 as vector, while DNA fragment 1417 bp is the protease gene insert. Therefore, the results of plasmid extraction from two clones were positively plasmid pBBRE194 prot-CM1.
The confirmed plasmid of pBBRE194 prot-CM1 was transformed to E. coli S17 as the donor cell for conjugation process. Escherichia coli S17 harboring pBBRE194 prot-CM1 was then conjugated to B. subtilis DB104 as the recipient cell. Conjugation to B. subtilis DB104 was obtained 20 clones that successfully grew on the selection media (calcium casein agar) containing erythromycin 5 µg/mL, but there was only one clone that produced a clear zone, that was clone 2. Clone 2 was tested for proteolytic activity by using control B. subtilis DB104 which harboring empty plasmid. The result showed that clone 2 produced a larger clear zone than control, with proteolytic index was 1.4 and control was 1.2 (Fig 3A).
The clone 2 of B. subtilis DB104 transconjugant was verified by using PCR method (Fig 3B). The result showed a DNA band of 1,417 bp located between 1,400-1,500 bp markers. The DNA band of 1 417 bp is the protease gene (insert) from B. halodurans CM1, because it was successfully amplified by using the same primer as when amplified insert to be ligated into vector pBBBRE194. Therefore, it was concluded that clone 2 was a positive clone carrying the recombinant plasmid pBBBRE194 prot-CM1.
Analysis of the expression of protease gene product in the recombinant B. subtilis DB104 was carried out by testing protease activity using crude enzyme extract. Protease activity was tested based on the “Amano” protease assay [21]. Protease enzyme activity of the recombinant B. subtilis DB104 was compared with B. subtilis DB104 in LB, skim milk 2%, and LB corn cobs 4% with the aim to determine the difference in the amount of enzyme produced. The protease enzyme activity from the recombinant B. subtilis DB104 had a higher activity than the protease enzyme produced by B. subtilis DB104 in both of production media. In LB with 2% skim milk, the recombinant B. subtilis DB104 had an activity of 4.26 U/ mL, while B. subtilis DB104 had an activity of 2.34 mL. Meanwhile, in the media of LB with 4% corn cobs, protease activity of the recombinant B. subtilis DB104 was 7.38 U/mL and B. subtilis DB104 was 5.42 U/mL (Fig 4A). The ability of the recombinant B. subtilis DB104 to produce a higher protease enzyme than B. subtilis DB104 proved that the protease gene in the plasmid pBBRE194 prot-CM1 can be expressed properly in the recombinant B. subtilis DB104
 |
Figure 4: Comparison of activity and protein concentration of protease produced by the recombinant B. subtilis DB104 and B. subtilis DB104 (control). |
Characterization of recombinant protease enzyme was carried out on enzyme produced by the recombinant B. subtilis DB104 in LB with 4% corn cobs. This is based on the consideration that the recombinant B. subtilis DB104 in LB with 4% corn cobs was obtained higher protease activity than LB with 2% skim milk. The recombinant protease enzyme was characterized based on the influence of temperature and pH, with temperature range of 30-60oC and pH range of 5-9.
This result showed that activity of the recombinant protease enzyme increased in proportion to the rise of temperature from 30-50 °C, but decreased at a temperature of 60 °C (Fig. 5A). The highest protease enzyme activity was obtained at 50°C at 13.66 U/mL, but decreased at 60 °C with an activity of 11.96 U/mL. It occurred because the temperature increasing who exceed the optimum temperature caused structural bonds in the enzyme would break [24]. At the maximum temperature, enzyme would denatured because the structure of the open protein and the non-polar group within the molecule became open out, the protein solubility in the polar water decreased, so that enzyme activity decreased [25].
The activity of the recombinant protease enzyme increased in proportion to the pH increasing. The highest activity of the protease enzyme was obtained at pH 9 of 13.66 U / mL, which proved that the protease enzyme produced by the recombinant B. subtilis DB104 worked best at alkaline pH (Fig 5B). Protease enzymes that are able to work in the range of neutral to alkaline pH (7-12) are grouped into alkaline proteases [26, 27]. The protease enzyme produced by B. subtilis DB104 recombinant has good activity at pH 7-9, so it belongs to the alkalthermophilic protease group.
 |
Figure 5: The effect of temperature (A) and pH (B) on the protease enzyme activity of the recombinant Bacillus subtilis DB104. |
The protein level in the recombinant protease enzyme from B. subtilis DB104 has a higher protein level than B. subtilis DB104 in both production media (LB with 2% skim milk and LB with 4% corn cob. Protein levels of the protease enzyme produced by B. subtilis DB104 recombinant and B. subtilis DB104 on LB with 4% corn cobs media were 0.464 mg/mL and 0.381 mg/mL, respectively. Meanwhile, the protein content of the protease enzyme produced by B. subtilis DB104 recombinant and B. subtilis DB104 in LB with 2% skim milk media was 0.434 and 0.341 mg/mL (Fig 4B). The recombinant B. subtilis DB104 and B. subtilis DB104 produced higher protein content in LB with 4% corn cobs than LB with 2% skim milk. This is because corn cobs are a good source of natural carbon for bacterial growth [28], so that protein levels can be produced in greater quantities.
Despite of the availability of whole gene information of Bacillus halodurans C-125 and the several report described properties of many other proteases from Bacillus genus [29, 30. 31], there was no report regarded to the cloning of protease gene from Bacillus halodurans in Bacillus subtilis. The isolation of ORF of this protease in E.coli has been also reported by our group [15], this study is the continued research and first report of cloning of B. halodurans protease gene in B. subtilis.
Conclusion
The protease gene from B. halodurans CM1 was successfully subcloned into the expression vector pBBRE194 and produced a recombinant plasmid pBBRE194 prot-CM1 with 9 819 bp size. Plasmid pBBRE194 prot-CM1 was successfully transformed by conjugation to B. subtilis DB104 and selected 1 positive clone that had been verified by plasmid and produced a clear zone. The recombinant protease enzyme produced by B. subtilis DB104 recombinant has higher activity than B. subtilis DB104, and the highest activity at 50 °C and pH 9 of 13.66 U/mL, so it is considered belonging to alkalothermophilic proteases.
Acknowledgement
This work was funded by INSINAS Research Incentives Program of Ministry of Research, Technology, and Higher Education in the year of 2018-2019 granted to IH.
References
- Selvam, K., M. Govarthanan, B. Shentilkumar, D. Senbagam, T. Selvankumar, S. Kamala-Kannan, & Shundakar. 2016. Optimization of protease production from surface modified coffee pulp waste and corncobs using Bacillus sp. by SSF 3. J. Biotechnol., 2016; 6: 167.
- Chu, W.H. 2007 Optimization of extracellular alkaline protease production from species of Bacillus . Ind. Microbiol. Biotechnol., 2007; 34: 241 —245.
- Gohel, S.D. & S.P. Singh. 2012. Cloning and expression of alkaline protease genes from two salt-tolerant alkaliphilic actinomycetes in Escherichia coli. Biol. Macromol., 2012; 50: 666—671.
- Naiola, E, Widhyastuti, N. Semi purifikasi dan karakterisasi enzim protease Bacillus Berkala Penelitian Hayati., 2007, 13: 51—56.
- Bhunia, B., B. Basak, Bhattacharya, T. Mandal, & A. Dey. Effect of pH and temperature on stability and kinetics of novel extracellular serine alkaline protease (70 kda). Int. J. Biol. Macromol., 2013; 54: 1—8.
- Kasana, R. Protease from psychrotrophs: An overview. J. Critical. Rev. Microbiol., 2010; 36: 134—145.
- Contesini, F.J., R.R. Melo, & H.H. Sato. An overview of Bacillus proteases: from production to application. J. Critical Rev. Biotechnol., 2017; 8: 1—
- Jiao, P., Q. Huang, & Y. Zhang. Gene cloning and expression of an alkaline serine protease with dehairing function from Bacillus pumilus. J. Curr. Microbiol., 2004; 49: 165—
- Shrinivas, D., R. Naik. Characterization of alkaline thermostable keratinolyitic protease froom thermoalkalophilic Bacillus halodurans JB 99 exhibiting dehairing activity. J Int. Biodet. Biodeg., 2010; 65(1): 29—35.
- Songyi, Zhang M., Liu, J., Jones G.S. Construction and application of recombinant strain for the production of an alkaline protease from Bacillus licheniformis. J. Biosci. Bioeng., 2014; 119(3): 284—288.
- Pakpahan, R. 2009. Isolasi bakteri dan uji aktivitas protease termofilik dari sumber air panas sipoholon tapanuli utara sumatera utara. [Tesis]. Universitas Sumatera Utara, Medan.
- Ulfah M., Helianti, I., Wahyuntari, , Nurhayanti, N. Characterization of a new thermoalkalophilic xylanase producing bacterial strain isolated from cimanggu hot spring, West Java, Indonesia. Microbiol. Indones., 2011; 5(3): 139—143.
- Takami, H., Horikoshi Reidentification of facultatively alkaliphilic Bacillus sp. C-125 to Bacillus halodurans. Biosci. Biotechnol. Biochem., 1999; 63(5): 943—945.
- Takami, H., Nakasone, , Takaki, Y., Maeno, R., Sasaki, N., Masui, Fuji, C., Hirama, Y. Nakamura, N., Ogaswara, S., Kuhara, Horikoshi, K. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans C-125 and genomic sequence comparison of Bacillus subtilis. Nuc. Acids. Res., 2000; 28(21): 4317—4331.
- Helianti,, Furgeva, N., Mulyawati, L., Ferniah, R.S., Kusumaningrum, H.P. Cloning of a gene encoding protease from Bacillus halodurans CM1 into Escherichia coli DH5a and expression analyses of the gene product. Makara J. Sci., 2018; 22(3):113-120
- Helianti, I., M. Ulfah, N. Nurhayati, & L. Mulyawati. Cloning, sequencing, and expression of the gene encoding a family 9 cellulase from Bacillus licheniformis F11 in Escherichia coli and Bacillus megaterium, and characterization of the recombinant enzymes. Indones., 2014; 8(4): 147—160.
- Sambrook, J. & D.W. Russel. Molecular cloning: a laboratory manual, 3rd2001; Cold Spring Harbor Laboratory Press, New York.
- Aquino de Muro, M. & F.G. Priest. Construction of chromosomal integrants of Bacillus sphaericus 2362 by conjugation with Escherichia coli. J. Microbiol., 2000; 151: 547—555.
- Richhardt J., Larsen M., & Meinhardt F. An improved transconjugation protocol for Bacillus megaterium facilitating a direct genetic knockout. Microbiol. Biotechnol., 2010; 86(6): 1959—1965.
- Schaeffer P., J. Millet, & J.P. Aubert. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA., 1965; 54:704—
- Amano, M. 2007. Amano enzyme. Nagoya, Japan: Amano EnzymeInc. Pp 77 —
- Ilmiah, S.N., Mubarik, R., Wahyuntari, B. Characterization of protease from Bacillus licheniformis F.11 as biodetergent agent. Makara J. Life. Sci., 2018; 22(3): 105—112.
- Bradford,M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem., 1976; 72(1—2): 248—254.
- Pratiwi,T. Mikrobiologi farmasi. Erlangga, Jakarta. 2008.
- Lehninger, A.L. Biochemistry. Akademic Press, New York.
- Gupta, R., Q.K. Beg, Lorenz, P. Bacterial alkaline protease molekular approach and industrial application. Microbiol. Biotechnology., 2002; 59: 5—32.
- Ellaiah, P., Srinivasulu, , Adinarayana, K. A Review on microbial alkaline proteases. J. Sci. Ind. Res., 2002; 61: 690—704.
- Erika, Agustrina, , Sumardi, Mulyono. Optimasi media produksi xilanase dari Bacillus sp. J. Selulosa., 2016; 6(1): 19—26.
- Hakim, A., Bhuiyan, F.R., Iqbal, A., Emon, T.H., Ahmed, J., Azad, A.K., Production and partial characterization of dehairing alkaline protease from Bacillus subtilis AKAL7 and Exiguobacterium indicum AKAL11 by using organic municipal solid wastes. , 2018; 4(6). doi: 10.1016/j.heliyon.2018.e00646.
- Marathe, S.K., Vashistht, M.A., Prashanth, A., Parveen, N., Chakraborty, S., Nair, S. Isolation, partial purification, biochemical characterization and detergent compatibility of alkaline protease produced by Bacillus subtilis, Alcaligenes faecalis, and Pseudomonas aeruginosa obtained from sea water samples. J. Genet. Eng. Biotechnol., 2018: 16(1):39-46. doi: 10.1016/j.jgeb.2017.10.001
- Singh, S., Gupta, P., Bajaj, B.K. Characterization of a robust serine protease from Bacillus subtilis K-1. J. Basic. , 2018; 58(1):88-98. doi: 10.1002/jobm.201700357.

This work is licensed under a Creative Commons Attribution 4.0 International License.