How to Cite | Publication History | PlumX Article Matrix
Faria Akbar, Susmita Saha,
Susmita Saha, Meghla Saha Pinky
Meghla Saha Pinky and Kazi Nahida Begum*
and Kazi Nahida Begum*
Department of Botany, Faculty of Life and Earth Sciences, Jagannath University, Chittaranjan Avenue, Dhaka-1100, Bangladesh
Corresponding Author E-mail: kazinahida@bot.jnu.ac.bd
DOI : http://dx.doi.org/10.13005/bbra/2879
ABSTRACT:
Brassica L. is the most agronomical important genus of Brassicaceae family. An electrophoretic exploration was conveyed for proper identification of genetically diverse and agronomically superior genotypes and pursuing the extent of genetic divergence and phylogenetic relationship within the thirteen variants of Brassica for leaf storage protein by using Polyacrylamide Gel Electrophoresis (PAGE) as biochemical marker. A total of 19 alternative protein bands were found with high polymorphism of 89.47%. The protein banding pattern suggested the existence of differences among the studied variants pertaining to the location, molecular weight and staining intensity of the bands which could be utilized as fingerprints for variants identification. Based on Nei’s genetic distance, a wide range of genetic distance (0.0541–1.5581) offered the presence of broad genetic variability among the quested variants. A dendrogram was constructed by using UPGMA where all the analyzed Brassica variants were rouped into two major clusters. Relying on this analysis, highest genetic variation (1.5581) was observed between BS-10 and BS-14 while the lowest genetic variation (0.0541) was recorded between BS-9 and BS-12, which might be furnished as a source of parental line. Consequently, it can be proposed that the protein profile of the analyzed thirteen variants of Brassica L. by PAGE would be considered to be a contributory implement to the breeders of Brassica by providing sufficient information on the genetic resources of Brassica and improvement of new offspring in the forthcoming breeding program of Brassica L.
KEYWORDS: Brassica L; Genetic diversity; Leaf protein; PAGE
Download this article as:| Copy the following to cite this article: Akbar F, Saha S, Pinky M. S, Begum K. N. Molecular Characterization of Thirteen Oil seed Brassica L. Variants From Bangladesh Through Polyacrylamide Gel Electrophoresis (PAGE). Biosci Biotech Res Asia 2020;17(4). |
| Copy the following to cite this URL: Akbar F, Saha S, Pinky M. S, Begum K. N. Molecular Characterization of Thirteen Oil seed Brassica L. Variants From Bangladesh Through Polyacrylamide Gel Electrophoresis (PAGE). Biosci Biotech Res Asia 2020;17(4). Available from: https://bit.ly/39G26yP |
Introduction
The Brassicaceae (Cruciferae) family consists of 338 genera and about 3709 species1. Among these genera, economically Brassica is the most important genus with 37 different species2. The genus – Brassica is composed of six interlinked species with great morphological and genetic diversity of which three diploid species- Brassica rapa (A genome), B. nigra (B genome), and B. oleracea (C genome) of the genus Brassica were considered to be responsible for the origin of three amphidiploid species, B. carinata (n = 17, BC genome), B. juncea (n = 18, AB genome) and B. napus (n = 19, AC genome)3. Brassica was originated near the Himalayan region. Brassica offered with a large number of important vegetables, oilseed and condiment crops also a great source of bioactive compounds, minerals, phytochemical contents, vitamins and fibers4.
Generally, the genus Brassica L. has been classified into three groups particularly –rapeseed, mustard and cole. The mustard groups include species like B. juncea, B. nigra and B. carinata; whereas the rapeseed groups include B. rapa and B. napus5. Commercial production of Brassica has grown progressively as a vital source of oil and plant originated protein for human and animal nutrition. Currently, rapeseed categorizes as the third source of vegetable oil (after soy and palm) and for oil meal it ranks as the third notable source (after soy and cotton)6. Brassica L. generates Indole-3-carbinol that plays significant role in reducing the growth of human breast cancer cells and the occurrence of tumors in reproductive organs7-8.
Moreover, Brassica L. are not only a quality sources of potassium, dietary fiber, phenolics, vitamins A, C and E but also use as a renewable resource or biofuel in the petro-chemical industry9. Higher protein solubility is found in seeds of B. napus than B. rapa seeds. As a rich source of edible protein, B. rapa, B. juncea, B. carinata, and B. nigra have commercial values in food industry. In oil-extraction process, rapeseed and canola meal remain as by-product which contain up to 42.7% to 50.00% protein10.
Evaluation with molecular marker facilitates in determining parental forms for mapping of population, marker assisted alternatives, line drawings of back crosses and consequently various molecular markers are applied to execute different studies which offer assistance to the breeders to improve crop species11. Now-a-days storage proteins are widely used as biochemical markers to find genetic structure, genetic diversity and relationships within plant species. According to O’Farrell12, polyacrylamide gel electrophoresis (PAGE) has been a well-accepted proteomic analytic method since its initiation to access protein banding patterns among different plant varieties. Hence electrophoresis of protein is considered as a method for characterization and evaluation of germplasm as well as increase the utilization of various plant genetic resources13.
Nowadays, storage protein is one of the most significant implements used to appraise genetic assortment among wild and cultivated plant species. Reviewing a number of earlier works of Mukhlesur and Hirata14, Sadia et al.15, Turi et al.13, Zada et al.16, Ibrahim et al.17 and so on, it has been revealed that they conveyed abundant efforts to find out genetic diversity and relationship among various species of Brassica for improvement of crop through SDS-PAGE.
Consequently the present investigation is conducted based on leaf storage protein of thirteen different BARI (Bangladesh Agriculture Research Institution) variants of Brassica from Bangladesh by utilizing PAGE technique to evaluate accurate protein profile for discerning variants, extent of genetic divergence and relationship among the thirteen inquired variants of Brassica as well as selection of parental line for further breeding program and crop improvement.
Materials and Methods
Plant Materials
Thirteen variants of the genus Brassica have been chosen for the current study reflecting a wide array of variation for diverse physio-morphological attributes (Table 1). To conduct the present investigation, all the thirteen variants of Brassica were collected from Oilseeds Research Center (ORC) of Bangladesh Agricultural Research Institute (BARI), Gazipur, Bangladesh and maintained in the Botanical garden of Jagannath University, Dhaka, Bangladesh. Analyses of leaf protein profile of the supplied variants were conducted in the laboratory of department of Botany, Jagannath University, Dhaka, Bangladesh.
Table 1: Some physio-morphological and agronomic traits of 13 Brassica L. variants used in the study.
| Species
|
Name of variants | Days to maturity | Siliquechamber | Seed color | Agronomic traits |
| B. juncea | Daulot (RS-81) | 90-105 | 2 chambered | Reddish brown | Comparatively long duration, low yielding variety. Tolerant to alternaria blight disease and environmental stresses like – drought and slightly tolerant salinity. |
| B. juncea | BARI Sarisha-2 (Rai-5) | 90-100 | 2 chambered | Reddish brown | Long duration, low yielding variety. Tolerant to drought, slightly tolerant to salinity. |
| B. juncea | BARI Sarisha-10 (BS-10) | 90-100 | 2 chambered | Reddish brown | Long duration, high yielding variety. Tolerant to drought, slightly tolerant to salinity. |
| B. juncea | BARI Sarisha-11 (BS-11) | 105-110 | 2 chambered | Reddish brown | Long duration, high yielding variety. Tolerant to drought and salinity. |
| B. napus | BARI Sarisha-7 (Napus-3142) | 90-95 | 2 chambered | Black | Long duration, high yielding variety, tolerant to alternaria blight disease and interim water logged condition. |
| B. rapa | BARI Sarisha-1 (Tori-7) | 70-80 | 2 chambered | Black | Short duration, low yielding variety. Susceptible to pest and diseases. |
| B. rapa | Kollaynia (TS-72) | 85-90 | 2 chambered | Blackish brown | Short duration, low yielding variety. Susceptible to pest and environmental stresses. |
| B. rapa | Sonali Sarisha (SS-75) | 90-100 | 4 chambered | Golden yellow | Long duration, high yielding variety. Susceptible to alternaria blight disease. |
| B. rapa | BARI Sarisha-6 (Dholi) | 90-100 | 2 chambered | Yellow | Long duration, high yielding variety. Susceptible to environmental stresses. |
| B. rapa | BARI Sarisha-9 (BS-9) | 80-85 | 2 chambered | Reddish brown | Short duration, high yielding variety. |
| B. rapa | BARI Sarisha-12 (BS-12) | 85-90 | 2 chambered | Reddish brown | Short duration, high yielding variety. |
| B. rapa | BARI Sarisha-14 (BS-14) | 75-80 | 2 chambered | Yellow | Short duration, high yielding variety. |
| B. rapa | BARI Sarisha-15 (BS-15) | 80-85 | 2 chambered | Yellow | Short duration, high yielding variety. |
Methods
Protein Isolation and Sample Preparation
Fresh and young leaves of one-month old seedlings of investigated thirteen variants of Brassica were collected to isolate the crude protein. Based on the methodology of Akbar et al.11 the collected leaves of Brassica variants were gently washed with distilled water and then with ethanol to clean the microspores and other dirt from the leaves surface and then kept on flitter papers for a while to soak up the excessive amount of distilled water and ethanol from the leaves. Afterwards, 1 gm fresh leaf of each sample was grinded and homogenized with cold deionised doubled distilled water in icy motor-pestle and later the crude homogenates were centrifuged at 4ºC with 13000 rpm for 15 min. After centrifugation, the crude protein remained as clear supernatant and stored in refrigerator at -20ºC as sample (isolated protein) for vertical polyacrylamide gel electrophoresis.
Estimation of Protein
According to Lowry et al.18, with little modification, the protein concentration of each investigated Brassica L. sample was estimated using 665 nm wave length via UV VIS spectrophotometer (UVN 15 Spectrophotometer).
Electrophoresis
Polyacrylamide gel electrophoresis of each inquired sample was conducted by following the strait of Akbar et al.11. The entire process of electrophoresis was performed by using omniPAGE mini vertical gel electrophoresis unit. The isolated protein sample was directly resolved with 10.0% polyacrylamide as separating gel and 4.0% as stacking gel during electrophoresis. Then, 200 μgm of each sample protein was loaded with 2X diluted Bromo Phenol Blue (BPB) loading dye (0.5 M Tris-HCl, pH 6.8; glycerol and deionised doubled distilled water in a volumetric ratio of 1: 1: 3 with a pinch of Bromo Phenol Blue) into the well of stacking gel. To run the electrophoresis, Cleaver nanoPAC– 300 constant power supply unit was employed and voltage was set at 90 V. The protein sample with BPB loading dye was mobilized in 10X diluted running buffer solution (Tris-glycine buffer, pH 8.3) until the dye front line arrived on 2mm above to the end of the gel. Afterwards, the gel was stained by 0.25% Coomassie Brilliant Blue (CBB) R-250 for 25 min and distained in acetic acid- methanol-distilled water (1: 4: 5 volume ratios) until the clear bands appeared on the gel. Lastly, the distained gel was gently washed with distilled water and the photographs of the gel were taken by a DSLR (18 mega pixels Canon EOS 700D model).
Data Analysis
Evaluations of the gels were done with bare eyes on a light box. The relative mobility (Rf values) of protein subunits were calculated by measuring the migration distance from the top of the separating gel to each band and to the dye front. For each band on the gel, the Rf value was calculated using the following equation:
Rf = migration distance of the protein / migration distance of the dye front
Determination of the discernible molecular weight of individual protein subunits was carried out using molecular weight marker proteins. Phosphorylase B, 97.2 KD; Bovine serum albumin, 66.4 KD; Ovalbumin, 44.3 KD; Carbonic anhydrase, 29.0 KD; Trypsin inhibitor, 20.1 KDa and Lysozyme, 14.3 KDa (Protein Molecular Weight Marker; TakaRa Bio USA) applied on the gel as marker protein. A standard curve of the log molecular weight (MW) on X axis versus relative mobility (Rf) of marker protein on Y axis was generated using computer based program Microsoft Excel. Molecular weight of individual unknown protein subunit from PAGE was determined by utilizing the equation:
y = mx + c, where y denotes for the molecular weight of unknown protein subunit.
The photographs of the gel were acutely reviewed on the basis of the presence (1) and absence (0) of protein bands. All the major and minor bands that were apparent to the eyes were considered in our current analysis and scoring of all the monomorphic and polymorphic bands were recorded. The scores acquired from PAGE analysis were then pooled for creating a single data matrix. Thereafter, the data was used to estimate proportion of polymorphic loci, Nei’s19 gene diversity (h), Shannon’s Information index20 and Nei’s21 genetic distance (D) employing a computer program, POPGENE (version 1.32)22. Based on genetic distance between all pairs of individual variants, a dendrogram was prepared applying Unweighted Pair Group Method with Arithmetic averages (UPGMA).
Results and Discussion
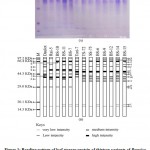
Proteins are thought about to be the forthright outcome of genes and might be used as markers of these genes. For characterizing systematic classification, protein might be performed as a supplemental mean. Hence an ample electrophoretic protein banding profile was conducted after the extraction and separation of stored leaf protein from the studied thirteen variants of Brassica L. through PAGE technique and represented in Fig. 1.
The electrophoretic protein banding patterns of the thirteen studied variants of Brassica were detected which conveyed to the marking off a total of nineteen polypeptide bands. Nineteen polypeptide bands were found to be present at nineteen different loci designated as a – s with molecular weight ranging from 16.36 to 97.20 KDa (Table 2 and Figs. 1A–B). A close inquisition of the bands displayed that the different variants had slight differences in their protein banding patterns with respect to the presence and absence and staining intensities of the bands. Moreover in the current study, the entire protein banding pattern of the investigated variants were grouped into 3 different regions (I to III) based upon the manner of increasing Rf values and decreasing molecular weight of proteins (Table 2).
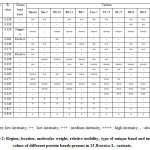 |
Table 2: Region, location, molecular weight, relative mobility, type of unique band and intensity values of different protein bands present in 13 Brassica L. variants. |
As a consequence, region – I was found to consist of six bands having a range of molecular weight 55.72–97.20 KDa with Rf value ranging from 0.028–0.173. The six protein bands of region – I was characterized with bands of mostly low intensity at loci – a, b, d and f, whereas in locus – e, moderate and low level intensity protein bands were found, protein bands of high intensity in all the variants were observed at locus – c, except in Rai-5 (negative unique band). The only low intensity protein band with molecular weight and Rf value of 72.95 KDa and 0.081 respectively, was detected from BS-10 at locus – d, which can be considered as a unique band (positive unique band) and may be used as an implement for particular varietal characterization of Brassica L. (Table 2 and Figs. 1A–B). All the bands in region–I was found to be polymorphic. Region – II was observed with nine protein bands having molecular weight and Rf value ranging from 38.46–52.76 KDa and 0.192–0.353, respectively. The protein bands revealed in this region were pre-eminently discerned with medium to high intensity of proteins at loci – m, n and o whose Rf value lie between 0.295–0.353. Two bands with low and high intensities of protein were present at loci – h and i. A single band with all low intensities of protein (at locus – j) and low and moderate intensities of protein were appeared at locus – l. At locus – g, a single band of low intensity protein with Rf value of 0.192 and molecular weight 52.76 KDa was apparent in Daulot whereas no bands of protein was observed in the rest of the twelve variants of Brassica at this locus, which made the band to be envisaged as an unique band (positive unique band) and assist in distinguishing the variant – Daulot from the other examined variants of Brassica (Table 2 and Figs. 1A–B). The region – III was characterized with very low intensities of protein bands found to be present at loci – p to s with molecular weight and Rf values ranging from 16.36–30.45 KDa and 0.501–0.854 (Table 2 and Figs. 1A–B). By taking into account the intensity of protein bands in different loci of the entire electrophoretic profile, it was observed that the region – II was more diverse with an average of 3 bands as compared to region – I and III, where the average numbers of bands were 2 and 1.33, respectively.
The change-over in the staining intensity and number of the polypeptide bands might be by virtue of differential extraction or disparity in solubility of protein or inadequacy of separation of varied sorts of proteins having identical migration rates23. They also suggested that the qualities of bands (i.e., the difference in the number, position and intensity of bands) in varieties even in accessions of the same species are governed through the quantitative gene system. Observations based on intensity of protein bands from different varieties of Brassica species were reported by many investigators13, 15, 24-25.Likewise, delineations on divergent plant species as regards to the intensities of protein bands was debriefed by Odeigah et al.26 from Nigerian varieties of pepper, Devi27 in sunflower, Varma et al.28 in maize genotypes, Vijayan29 in rice, Paul and Datta30 in celery and ajowan, Nisha31 in wheat, Sumathi32 in oats, Abdulrahaman et al.33 in lady’s finger and Begum and Alam34 in chick-pea.
Divergence within the loci concerning the position, staining intensity and values of molecular weight were observed in the electrophorogram (Table 2 and Figs. 1A–B). Two bands with molecular weight 55.72 and 49.25 KDa (one low and another one was high in intensity) were obtained at loci – f and i, respectively which were found to be present in all the inquired variants of B. rapa (Tori-7, TS-72, SS-75, BS-6, BS-9, BS-12, BS-14 and BS-15) whereas two consecutive bands of very low intensity with molecular weight of 30.45 and 27.89 KDa at loci – p and q were spotted out from the variants of B. juncea and B. napus (Daulot, Rai-5, BS-10, BS-11 and BS-7). The presence of bands with particular molecular weight at definite locus within particular species made us to imply that the species B. rapa exhibited species specific bands at loci f and I of molecular weight 55.72 and 49.25 KDa. Concurrently, B. juncea and B. napus displayed species specificity for molecular weight 30.45 and 27.89 KDa at p and q loci, discretely. Thereupon, in the light of diversity regarding to the position, intensity and values of molecular weight for each of the locus can be aided as feasible tool for proper distinguishing of species within the observed variants of Brassica by the electrophoresis of the total soluble protein from leaves.
In our present inquest, scrutiny of soluble protein banding pattern from leaves of thirteen tested variants of Brassica L. by PAGE technique presented three distinct different profiles and could be an excellent genre of biochemical fingerprint for discerning different variants of Brassica. The presence of a negative (-ve) unique band at locus – c in Rai-5 and a positive (+ve) unique band in each of the variety of BS-10 and Daulot at locus – d and g singly, could be esteemed as fingerprints for discerning these respective variants (Figs. 1A–B and Table 2).
Inter varietal locus variation can be deemed to be a mainspring for estimation of different degree of genetic divergence within diverse species, where non – appearances of a few protein polypeptides in few variants express variation and consequently taken into considered as polymorphic loci. Throughout the course of the prevailing analysis, inter varietal variation of loci among the thirteen variants of Brassica L. were also disclosed and presented in Table 3. Out of total nineteen loci; the loci – m and s are vitally monomorphic because of the prevalence of 100% protein bands. The remaining loci of the entire electrophorogram exhibited variation within themselves. The highest amount of variation was observed from loci – d and g with 92.31% of variability and 0.08 genetic disagreement, whereas the lowest amount of divergence was noticed from locus – c with 7.69% variability that exists of a high value of genetic disagreement (0.92) (Table 3).
Table 3: Inter varietal locus variation among the 13 variants of Brassica L.
| Locus | Present (%) | Absent (%) | Variation (%) | Status | Genetic disagreement |
| Locus a | 11 (84.62%) | 2 (15.38%) | 15.38% | Polymorphic | 0.85 |
| Locus b | 4 (30.77%) | 9 (69.23%) | 69.23% | Polymorphic | 0.31 |
| Locus c | 12 (92.31%) | 1 (7.69%) | 7.69% | Polymorphic | 0.92 |
| Locus d | 1 (7.69%) | 12 (92.31%) | 92.31% | Polymorphic | 0.08 |
| Locus e | 10 (76.92%) | 3 (23.08%) | 23.08% | Polymorphic | 0.77 |
| Locus f | 9 (69.23%) | 4 (30.77%) | 30.77% | Polymorphic | 0.69 |
| Locus g | 1 (7.69%) | 12 (92.31%) | 92.31% | Polymorphic | 0.08 |
| Locus h | 6 (46.15%) | 7 (53.85%) | 53.85% | Polymorphic | 0.46 |
| Locus i | 11 (84.62%) | 2 (15.38%) | 15.38% | Polymorphic | 0.85 |
| Locus j | 6 (46.15%) | 7 (53.85%) | 53.85% | Polymorphic | 0.46 |
| Locus k | 8 (61.54%) | 5 (38.46%) | 38.46% | Polymorphic | 0.62 |
| Locus l | 4 (30.77%) | 9 (69.23%) | 69.23% | Polymorphic | 0.31 |
| Locus m | 13 (100.00%) | 0 (0.00%) | 00.00% | Monomorphic | 1.00 |
| Locus n | 9 (69.23%) | 4 (30.77%) | 30.77% | Polymorphic | 0.69 |
| Locus o | 7 (53.85%) | 6 (46.15%) | 46.15% | Polymorphic | 0.54 |
| Locus p | 5 (38.46%) | 8 (61.54%) | 61.54% | Polymorphic | 0.38 |
| Locus q | 5 (38.46%) | 8 (61.54%) | 61.54% | Polymorphic | 0.38 |
| Locus r | 7 (53.85%) | 6 (46.15%) | 46.15% | Polymorphic | 0.54 |
| Locus s | 13 (100.00%) | 0 (0.00%) | 00.00% | Monomorphic | 1.00 |
| Total Polymorphism 89.47% | |||||
Besides, the showing up of different level of variableness in the loci of the studied variants like 15.38% (at loci – a and i), 23.08% (at locus – e), 30.77% (at loci – f and n), 38.46% (at locus – k), 46.15% (at loci – o and r), 53.85% (at loci – h and j), 61.54% (at loci – p and q) and 69.23% (at loci – b and l) along with respective genetic disagreement of 0.85, 0.77, 0.69, 0.62, 0.54, 0.46, 0.38 and 0.31 made it pondered to have worth mentionable genetic diversity among the quested varieties of Brassica L.
The proficiency of molecular marker approaches relies on the level of polymorphism within a group of inquired variants. During the study of PAGE, the nineteen bands of polypeptide were amplified of which seventeen (89.47%) were found to be polymorphic and the remaining two bands (10.53%) were monomorphic in nature (Fig. 2A). An average of 44.13% polymorphism was observed from the tested variants of Brassica L. (Table 4). The presence of high amount of polymorphism among the variants expressed by the proportion of polymorphic loci (89.47%), it can be suggested that a broad genetic variation may be present among the studied variants of Brassica L. From the outcome of the current study, the highest amount of polymorphism was documented from BS-10, which was 57.89%. As against BS-14 with 31.58% of polymorphism which was found as the lowest value of polymorphism among the examined variants of Brassica L. The studied variants of Brassica L. exhibited different degree of polymorphism displayed in Table 4 and Fig. 2B. Of the three investigated species of Brassica L., 42.11% and 42.76% of average polymorphism was revealed from B. napus and B. rapa, respectively whereas B. juncea was observed with 47.37% of average polymorphism (Table 4).
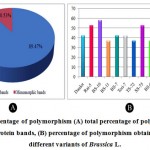 |
Figure 2: Percentage of polymorphism (A) total percentage of polymorphic and monomorphic protein bands, (B) percentage of polymorphism obtained from thirteen different variants of Brassica L. |
Table 4: Levels of polymorphism within 13 variants of Brassica L.
| Variants of Brassica
|
No. of polymorphic bands | % of polymorphism in Brassica variants | Average % of polymorphism in each Brassica species | Average % of polymorphism in 13 Brassica variants |
| Brassica juncea var. Daulot | 8 | 42.11 | 47.37 | |
| B. junceavar. Rai-5 | 10 | 52.63 | ||
| B. juncea var. BS-10 | 11 | 57.89 | ||
| B. juncea var. BS-11 | 7 | 36.84 | ||
| B. napus var. BS-7 | 8 | 42.11 | 42.11 | 44.13 |
| B. rapa var. Tori-7 | 8 | 42.11 | 42.76 | |
| B. rapa var. TS-72 | 7 | 36.84 | ||
| B. rapa var. SS-75 | 10 | 52.63 | ||
| B. rapa var. BS-6 | 7 | 36.84 | ||
| B. rapa var. BS-9 | 9 | 47.37 | ||
| B. rapa var. BS-12 | 10 | 52.63 | ||
| B. rapa var. BS-14 | 6 | 31.58 | ||
| B. rapa var. BS-15 | 8 | 42.11 |
As stated through Majumder et al.35 assessment of genetic diversity aided with protein markers have been ascertained as a sterling tool in characterization of many crops species at gene level. The values of Nei’s19 gene diversity and Shannon’s information index20 for the inquired Brassica variants across all the loci are provided in Table 5 and Fig. 3. The estimation of Nei’s19 genetic diversity for all the variants was 0.3488 ± 0.1726 and Shannon’s information index was 0.5098 ± 0.2299. Estimation of Nei’s19 gene diversity (0.3488) and Shannon’s information index (0.5098) across all loci (Table 5) also assisted with the subsistence of high level of genetic variation in all studied materials of Brassica L.
Table 5: Estimation of genetic variability among 13 variants of Brassica L.
|
Locus |
Nei’s (1973) gene diversity ( h ) |
Shannon’s information index ( i ) |
| Locus a | 0.3550 | 0.5402 |
| Locus b | 0.4260 | 0.6172 |
| Locus c | 0.1420 | 0.2712 |
| Locus d | 0.1420 | 0.2712 |
| Locus e | 0.3550 | 0.5402 |
| Locus f | 0.4260 | 0.6172 |
| Locus g | 0.1420 | 0.2712 |
| Locus h | 0.4970 | 0.6902 |
| Locus i | 0.4970 | 0.5402 |
| Locus j | 0.3550 | 0.6902 |
| Locus k | 0.4970 | 0.6902 |
| Locus l | 0.4260 | 0.6172 |
| Locus m | 0.0000 | 0.0000 |
| Locus n | 0.4260 | 0.6172 |
| Locus o | 0.4970 | 0.6902 |
| Locus p | 0.4794 | 0.6663 |
| Locus q | 0.4794 | 0.6663 |
| Locus r | 0.4970 | 0.6902 |
| Locus s | 0.0000 | 0.0000 |
| Mean | 0.3488 | 0.5098 |
| St. Dev | 0.1726 | 0.2299 |
 |
Figure 3: Estimated genetic diversity (Nei’s genetic diversity, Shannon’s information index and proportion of polymorphic loci found at different loci) of studied Brassica variants. |
The only disclosure on PAGE for leaf stored protein of Brassica L. was reported by Akbar et al.11, in which an average of 20.64% polymorphism was documented from three different varieties of the species – B. rapa, B. juncea and B. napus. Hence it can be suggested that more number of varieties and species are required to assess the degree of polymorphism of Brassica L. The determination of our existing perusal has been then assimilated with the findings of Mukhlesur and Hirata14, where the cultivars of B. rapa, B. juncea, B. napus, B. carinata, B. oleracea and hexaploid Brassica from various geographical origins were examined for leaf protein analysis by SDS-PAGE and the consequence of their analysis found incongruous with the outcome of the current study because they observed no significant difference within the examined cultivars even between different species of Brassica. The possible reasons for such type of inconsistency may be due to – (i) difference in morphology, ploidy level and constituents of genome, (ii) for different geographical distribution of the respective variants, (iii) difference in cultural practices, (iv) difference in methodological approaches.
Mentionable amount of studies have been performed early by many workers concerning the degree of polymorphism for total seed storage protein of Brassica as well as other species of cereal, pulses and oilseed crops with SDS-PAGE technique throughout the world. 21.20% of polymorphism was recorded within the varieties of B. campestris imitated by 6.30% in B. napus and 3.20% in B. juncea after evaluating varied varieties of distinctive Brassica species (Mukhlesur and Hirata)14. Ibrahim et al.17 apprised 58.00% of polymorphism from 53 genotypes of Indian mustard (B. juncea L.) germplasm. 94.44% polymorphism was reported from the diverse genotypes of Eruca sativa by Shinwari et al.36. From six cultivars of Egyptian soybean, a total of 30.43% polymorphism was assayed by Rayan and Osman37, 63.2% polymorphism was conveyed by Hlozáková et al.38 in four European cultivars of common wheat, in hundred-five accessions of Pakistani sesame 70.00% polymorphism was debriefed by Akbar et al.39, 82.00% of polymorphism have been evaluated by Vivodík et al.40 from fifty-six genotypes of Tunisian castor bean. Bhargav et al.41 assessed 91.00% of polymorphism from twenty Indian local genotype of common bean.
Knowledge on genetic similarity (distance) between germplasm and among individuals or populations is beneficial in an exceedingly breeding application since it lets incorporation of germplasm and offers greater effective sampling of germplasm to go for the improvement of populations. In the current research, the dendrogram constructed from the UPGMA analysis and coefficients of distance matrix unconcealed great connections between a numbers of variants (Fig. 4 and Table 6). By taking into account the banding pattern of leaf protein, genetic distance matrix for all the thirteen quested samples of Brassica variants were determined concurring to Nei’s21 genetic distance (Table 6).
Table 6: Summary of Nei’s genetic distances of 13 variants of Brassica L.
| Brassica variants | Daulot | Rai-5 | BS-10 | BS-11 | BS-7 | Tori-7 | TS-72 | SS-75 | BS-6 | BS-9 | BS-12 | BS-14 | BS-15 |
| Daulot | 0 | ||||||||||||
| Rai-5 | 0.3795 | 0 | |||||||||||
| BS-10 | 0.4595 | 0.3054 | 0 | ||||||||||
| BS-11 | 0.4595 | 0.1719 | 0.3795 | 0 | |||||||||
| BS-7 | 0.3795 | 0.2364 | 0.3054 | 0.0541 | 0 | ||||||||
| Tori-7 | 0.5465 | 0.5465 | 0.6419 | 0.3054 | 0.2364 | 0 | |||||||
| TS-72 | 0.8650 | 0.8650 | 0.9985 | 0.5465 | 0.6419 | 0.3054 | 0 | ||||||
| SS-75 | 0.7472 | 0.9985 | 0.8650 | 0.8650 | 0.9985 | 0.5465 | 0.1719 | 0 | |||||
| BS-6 | 0.8650 | 0.8650 | 1.3350 | 0.5465 | 0.6419 | 0.3054 | 0.2364 | 0.3054 | 0 | ||||
| BS-9 | 0.6419 | 0.6419 | 0.9985 | 0.5465 | 0.6419 | 0.4595 | 0.3795 | 0.3054 | 0.1112 | 0 | |||
| BS-12 | 0.7472 | 0.7472 | 0.8650 | 0.6419 | 0.7472 | 0.5465 | 0.4595 | 0.2364 | 0.1719 | 0.0541 | 0 | ||
| BS-14 | 0.7472 | 0.9985 | 1.5581 | 0.6419 | 0.7472 | 0.3795 | 0.1719 | 0.2364 | 0.0541 | 0.1719 | 0.2364 | 0 | |
| BS-15 | 0.7472 | 0.7472 | 1.1527 | 0.4595 | 0.5465 | 0.2364 | 0.1719 | 0.2364 | 0.0541 | 0.1719 | 0.2364 | 0.1112 | 0 |
Genetic variation among the variants typically screen via the way of means of genetic distance matrix. In our existing disclosure, the values of pair-wise comparison of Nei’s21 genetic distance among thirteen Brassica variants ranged from 0.0541 to 1.5581 (Table 6 and Fig. 4). The highest genetic distance was observed between BS-10 and BS-14 (1.5581) among the variants (Table 6) that indubitably demonstrated the presences of greater genetic distance between these two populations, notably the previous one representing the variant of B. juncea whilst the last mentioned one represented the variant of B. rapa. Likewise, pair wise genetic distance with relatively high values was detected between BS-10 and BS-6 (1.3350), BS-10 and BS-15 (1.1527), BS-10 and TS-72 (0.9985), BS-10 and BS-9 (0.9985), BS-7 and SS-75 (0.9985), Rai-5 and SS-75 (0.9985), Rai-5 and BS-14 (0.9985). Contrastingly, the lowest genetic distance was found between BS-9 and BS-12 (0.0541), both were variants from B. rapa (Table 6). The difference between the highest (1.5581) and the lowest value of genetic distance (0.0541) revealed the wide range of genetic variability persisting among the thirteen rapeseed-mustard variants. High genetic distance values between variants pair may be found because of difference in hereditary constituents. Taking account the genetic distance values, the findings revealed that variants were genetically distinctive from each other and which may well be utilized in breeding program to achieve potential hereditary picks up.
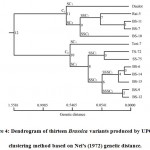 |
Figure 4: Dendrogram of thirteen Brassica variants produced by UPGMA clustering method based on Nei’s (1972) genetic distance. |
Based on Nei’s21 genetic distance obtained from protein banding pattern, a dendrogram was drawn up employing UPGMA in which the thirteen variants of Brassica were differentiated into two main clusters or groups C1 and C2 (Fig. 4). The first cluster C1 consisted of five variants of Brassica of which four variants from B. juncea (Daulot, Rai-5, BS-10 and BS-11) and one variant from B. napus, BS-7 were present. The explanation for two species (B. juncea and B. napus) had a place to same cluster is probable that the nearly introduced genotype might be shared some genes from the AA genomic base of the two variants which have been considered in this study. It might also possibly that the alleles of CC genome of the species (B. napus) may be near to that of the other elemental variants of the same cluster. The cluster or group C1 was divided into two sub-clusters. Daulot was found to be present in sub-cluster I (SC1) of cluster C1, and sub-cluster II (SC2) was further divided into two sub-sub cluster where sub-sub cluster II (SSC2) was found with only BS-10. Concurrently, sub-sub cluster I (SSC1) was consisted of three closely related variants of Rai-5 with BS-11 and BS-7. On the other hand, the major cluster C2 included eight variants of B. rapa (Tori-7, TS-72, SS-75, BS-6, BS-9, BS-12, BS-14 and BS-15) and divided into two sub-clusters (SC1) and (SC2). The sub-cluster (SC1) of cluster C2 was presented with Tori-7 alone; however the second sub-clusters (SC2) included the rest of the variants of B. rapa. The second sub-clusters (SC2) further segregated into two sub-sub clusters; SSC1 and SSC2. In sub-sub cluster I (SSC1), there was TS-72 and SS-75 whereas sub-sub cluster II (SSC2), was further divided in two sub-sub-sub clusters. Sub-sub-sub cluster I was present with three closely related variants of BS-6, BS-14 and BS-15 whereas, in sub-sub-sub cluster II there was BS-9 and BS-12 with minimal genetic distance of 0.0541. It was observed that the five variants of B. rapa found in sub-sub-sub cluster II showed low genetic distances among them ranging from 0.0541–0.2364. In this way, there was a limpid clustering pattern of geographically closer variants within the present ponder showing that the affiliation between genetic relatedness and geological distance has significance.
Presently, after the comparison between the come out of physio-morphological and agronomic traits and genetic distance matrix (Table 1 and Fig. 4) of the inquired variants of Brassica, it was revealed that in the first cluster C1, Daulot, Rai-5, BS-10, BS-11 and BS-7 had almost the similar seed color (reddish brown to black seed) and other agronomic characteristics (such as – tolerance to biotic and aboitic stresses of environment). Similarly, in the major cluster C2 – the morphological traits which include seed coloring were alike in BS-6, BS-14 and BS-15 (yellow seeded variants), whereas in BS-9 and BS-12 (reddish brown seeded variants). It was also observed that SS-75 and TS-72 grouped in a distant pair based on agronomic aspects (susceptibility to pest and environmental factors) and deviated from black seeded variety Tori-7 via cluster analysis (Fig. 4 and Table 1). The report of Saha et al.42 on genetic assessment of four Brassica species through RAPD marker found more or less congruent with our current disclosure that yellow seeded Brassica variants could be separated from the brown seeded variants by cluster analysis. Therefore, it looks through that cluster analysis would play a noteworthy implement in ascertaining genetic diversity regarding the diverse physio-morphological characters and other agronomic traits of plant species.
Conclusion
Genetic variation alludes to the differences within the constitutions of heredity in an individual of a species and it is imperative in keeping up the developmental steadiness and biological latent of plant species. More genetic variability within variants and noteworthy differentiation between variants states plenty of genetic resources of a species. The outcome of our existing quest exhibited differences in the position, number and staining intensity of protein bands among the studied variants which manifests the application of PAGE for differentiation of inquired variants of Brassica. High level of polymorphisms (89.47%) along with wide range of genetic distance (0.0541–1.5581) was viewed from the thirteen variants of Brassica. Broad range of polymorphism and genetic distance brought to light the presence of wide variability within Brassica spp. The variants of BS-10 and BS-14 contain the highest genetic variation, whereas BS-9 and BS-12 contain the lowest genetic variation among the variants employed in this analysis. Variants having near vicinity in their origin, morphological traits and stratagem of breeding are possibly to have less genetic distance from each other. Hence, the results of this inquisition put forward for consideration that the variants of BS-10, BS-9, BS-12 and BS-14 could furnished the amenities for selection as parental source in coming breeding program to ameliorate Brassica variants in Bangladesh. However, it is recommended that exceeding molecular information is required to own better evaluation of genetic variability of Brassica germplasm in Bangladesh as well as currently launched varieties/ lines and therefore more efficacious utilization of existing variability for advancement of Brassica crop in Bangladesh.
Acknowledgement
The authors expressed their gratitude to the Oilseed Research Center of Bangladesh Agricultural Research Institute (BARI) for providing the seeds of thirteen variants of Brassica L.
Conflict of Interest
The authors pertaining to this research work declared no conflict of interest.
Funding Source
Ministry of Science and Technology, Government of the People’s Republic of Bangladesh granted the financial support for conducting this research work.
References
- Kasem W. T., Ghareeb A., Marwa E. Seed morphology and seed coat sculpturing of 32 taxa of family Brassicaceae. Am. Sci. 2011; 7(2): 166–178.
- Gomez – Campo C. Studies on Cruciferae V. Chromosome numbers for twenty-five taxa. Anales del Instituto Botanica A. J. Cavanilles. 1980; 35: 177–182.
- Nagahara U. Genome analysis in Brassica with special reference to the experimental formation of napus and peculiar mode of fertilization. Jpn. J. Bot. 1935; 7: 389–452.
- Hedge I. C.: A systematic and geographical survey of old world Cruciferae. In: The biology and chemistry of the Cruciferae (Vaughan JG, MacLeod AJ, Jones BMG, eds). New York: Academic Press. 1976; pp 1–46.
- Yarnell S. H. Cytogenetics of the vegetable crops. II. Crucifers. Rev. 1956; 22: 81–166.
CrossRef - Thiyam U., Kuhlmann A., Stockmann H., Schwarz K. Prospects of rapeseed oil by-products with respect to antioxidative potential. R. Chim. 2004; 7(6): 611–616.
CrossRef - Cover C. M., Hsieh S. J., Tran S. H., Hallden G., Kim G. S., Bjeldanes L. F., Firestone G. L. Indole-3-carbonyl inhibits the expression of cyclin-dependent kinase-6 and induces a G1-cell cycle arrest of human breast cancer cells independent of estrogen receptor signaling. J. Biol. Chem. 1998; 273(7): 3838–3847.
CrossRef - Staub R. E., Feng C., Onisko B., Bailey G. S., Firestone G. L., Bjeldanes, L. F. Fate of indole-3-carbinol in cultured human breast tumor cells. Res. Toxicol. 2002; 15(2): 101–109.
CrossRef - Zhang J., Yuan Q., Meng Y., Li X., Nan Z., Wang Y., Zhang W. A genetic diversity analysis of wild Lespedeza populations based on morphological characters, allozyme and RAPD methods. Plant Breed. 2007; 126: 89–94.
CrossRef - Ghodsvali A., Khodaparast M. H. H., Vosoughi M., Diosady L. L. Preparation of canola protein materials using membrane technology and evaluation of meals functional properties. Food Res. Int. 2005; 38(2): 223–231.
CrossRef - Akbar F., Paul M., Begum K. N. Karyological and genetic diversity study using molecular marker among three species of oilseed Brassica Ind. J. Pure App. Biosci. 2020; 8(4): 267–281.
CrossRef - O’Farrell P. H. High resolution two-dimensional electrophoresis of proteins. Biol. Chem. 1975; 250(10): 4007–4021.
CrossRef - Turi N. A., Farhatullah, Rabbani M. A., Khan N. U., Akmal M., Pervaiz Z. H., Aslam, M. U. Study of total seed storage protein in indigenous Brassica species based on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). J. Biotechnol. 2010; 9(45): 7595–7602.
- Mukhlesur R. Md., Hirata Y. Genetic diversity in Brassica species using SDS-PAGE analysis. Biol. Sci. 2004; 4(2): 234–238.
CrossRef - Sadia M., Malik S. A., Rabbani M. A., Pearce S. R. Electrophoretic characterization and the relationship between some Brassica Electron. J. Biol. 2009; 5(1): 1–4.
- Zada M., Zakir N., Rabbani Z., Shinwari Z. K. Assessment of genetic variation in Ethopian mustard (Brassica carinata Braun) germplasm using multivariate techniques. Pak. J. Bot. 2013; 45: 583–593.
- Ibrahim M. I., Abbasi F. M., Rabbani M. A., Inamullah, Jan S. A., Ilyas M., Khurshid H. Evaluation of genetic variation among Indian Mustard (Brassica juncea) genotypes by SDS-PAGE method. Proc. Pakistan Acad. Sci. (B. Life and Environmental Sciences) 2017; 54(4): 333–339.
- Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. Biol. Chem. 1951; 193(1): 265–275.
CrossRef - Nei M. Analysis of gene diversity in subdivided populations. Natl. Acad. Sci. 1973; 70(12): 3321–3323.
CrossRef - Lewontin, R. C.: The apportionment of human diversity. In: Evolutionary Biology (Dobzhansky T, Hecht MK, Steere WC, eds). Heidelberg: Springer. 1972; pp 381–398.
CrossRef - Nei M. Genetic distance between populations. Nat. 1972; 106(949): 283–292.
CrossRef - Yeh F. C., Yang R. C., Boyle T. B. J., Ye Z. H., Mao J. X. POPGENE, the user-friendly software for population genetic analysis. Biol. Biotechnol. Cent. 1999; University of Alberta, Canada.
- Ladizinsky G., Hymowitz D. Seed protein electrophoresis in taxonomic and evolutionary studies. Appl. Genet. 1976; 54: 145–151.
CrossRef - Geetha V. V., Balamurugan P. SDS PAGE electrophoresis in mustard cultivars. J. Agric. Res. 2011; 6(5): 437–443.
CrossRef - Kakaei M., Kahrizi D. Study of seed proteins pattern of Brassica napus varieties via sodium dodecyl sulfate polyacrylamid gel electrophoresis. Res. J. Biotechnol. 2011; 2(1): 26–28.
- Odeigah P. G. C., Oboh B., Aghalokpe I. O. The characterization of Nigerian varieties of pepper, Capsicum annuum and Capsicum frutescens by SDS-polyacrylamide gel electrophoresis of seed proteins. Resour. Crop Evol. 1999; 46: 127–131.
CrossRef - Devi S. G. Varietal identification through electrophoresis in sunflower (Helianthus annus) 2000; Master’s thesis, Tamil Nadu Agricultural University, Coimbatore.
- Varma V. L. K., Reddy N. M., Keshavulu K., Ankaiah R. Characterization of maize (Zea mays) genotypes through seed protein electrophoresis (SDS-PAGE). Crop. Res. 2005; 30: 124–127.
- Vijayan R. Organic seed production in rice cv. ADT 43. 2005; Doctoral dissertation, Tamil Nadu Agricultural University, Coimbatore.
- Paul R., Datta A. K. Seed protein profiles in celery (Apium graveolens) and Ajowan (Trachyospermum amni L.) plant types. Int. J. Plant. Sci. 2006; 1(1): 36–38.
- Nisha C. Assessment of seed quality in wheat (Triticum aestivum L.) varieties as influenced by different alien rust resistant genes under varying production conditions. 2007; Doctoral dissertation, Tamil Nadu Agricultural University, Coimbatore.
- Sumathi S. Varietal identification in oats (Avena sativa ). 2007; Master’s thesis, Tamil Nadu Agricultural University, Coimbatore.
- Abdulrahaman A. A, Kolawole O. S., Onile O. G., Oladele F. Seed electrophorentic characterization and taxonomic implications of some accessions of Abelmoschus esculentus (Moench) in Nigeria. Jewel J. Sci. Res. 2015; 3(1): 136–145.
- Begum K. N., Alam Sk. S. Genetic diversity in nine chick-pea (Cicer arietinum) varieties based on different molecular markers. Bangladesh J. Bot. 2019; 48(1): 195–203.
CrossRef - Majumder D. A. N., Hassan L., Rahim M. A., Kabir M. A. Analysis of genetic diversity in mango (Mangifera indica) using isozymetic polymorphism. Afr. J. Biotechnol. 2012; 11(87): 15310–15323.
- Shinwari S., Akbar F., Rabbani M. A., Mumtaz A. S., Shinwari Z. K. Evaluation of genetic diversity in different genotypes of Eruca sativa from Pakistan by SDS-PGE analysis. J. Bot. 2013; 45(4): 1235–1240.
- Rayan W. A., Osman S. A. (2019). Phylogenetic relationships of some Egyptian soybean cultivars (Glycine max) using SCoT marker and protein pattern. Bull. Natl. Res. Cent. 2019; 43(161).
CrossRef - Hlozáková T. K., Gregová E., Vivodík M., Gálová, Z. Genetic diversity of European cultivars of common wheat (Triticum aestivum) based on RAPD and protein markers. J. Cent. Eur. Agric. 2016; 17(4): 957–969.
CrossRef - Akbar F., Yousaf N., Rabbani M. A., Shinwari Z. K., Masood M. S. Study of total seed proteins pattern of sesame (Sesamum indicum) Landraces via sodium dodecyl sulfate polyacrylamide gel elecrophoresis (SDS-PAGE). Pak. J. Bot. 2008; 44(6): 2009–2014.
- Vivodík M., Saadaoui E., Balážová Ž., Gálová Z., Petrovičová L. Characterization of Tunisian castor bean genotypes using SDS-PAGE of total seed storage proteins. S. J. F. Sci. 2018; 12(1): 701–706.
CrossRef - Bhargav S., Rai G. K., Mallick S. A., Rai S. K., Kumar R. R., Singh M., Parveen A., Jamwal D., Saikia S. L., Bagati S. Assessment of genetic diversity in local genotypes of common bean (Phaseolus vulgaris) using biochemical markers. The Bioscan 2016; 11(3): 1955–1961
- Saha S., Molla Md. R., Chandra D., Rahman, L. Assessment of genetic variation and relationships within the varieties of four Brassica species by RAPD markers. J. Crop Sci. 2008; 2(3): 105–114.

This work is licensed under a Creative Commons Attribution 4.0 International License.