How to Cite | Publication History | PlumX Article Matrix
Jyotish Sonowal, Pankaj Chetia and Devid Kardong
Department of Life Sciences, Dibrugarh University, Dibrugarh, Assam, India
Corresponding Author E-mail: kardongdevid@dibru.ac.in
DOI : http://dx.doi.org/10.13005/bbra/2907
ABSTRACT:
To update the present knowledge of freshwater molluscs, a phylogenetic analysis of two species of Indian pond mussels was carried out using amplified sequence of 18S rRNA gene. From the phylogenetic study of 18S rRNA gene sequencing, it was found that the Lamellidenscorrianus and L. phenchooganjensis are phylogenetically closely related to 18S rRNA gene sequences of other freshwater bivalve mussels belonging to the family Unionidae of order Unionida. Like other members of the Unionidae family, the two Lamellidensspp. showed monophyletic evolutionary lineage and shared a common ancestor. The result obtained from the phylogenetic analysis of Lamellidensspp. was significant as the 18S rRNA gene sequence of L. phenchooganjensis was submitted for the first time in the global nucleotide database (Genbank, NCBI). Similarly, the 18S rRNA gene sequence of L. corrianuswas also submitted to the database for the first time from this region having unique ecological niche. Therefore, the present study on phylogenetic analysis is a contribution to the global database of 18S rRNA gene sequences of freshwater mollusc, particularly from this part of the region.
KEYWORDS: Common Ancestor; Database; Data Deficient; Monophyletic; Unionidae
Download this article as:| Copy the following to cite this article: Sonowal J, Chetia P, Kardong D. Phylogenetic analysis of Indian freshwater pond mussels Lamellidenscorrianus and L. phenchooganjensis (Bivalvia: Unionidae) from the upper Brahmaputra Basin of Assam, India. Biosci Biotech Res Asia 2020;18(1). |
| Copy the following to cite this URL: Sonowal J, Chetia P, Kardong D. Phylogenetic analysis of Indian freshwater pond mussels Lamellidenscorrianus and L. phenchooganjensis (Bivalvia: Unionidae) from the upper Brahmaputra Basin of Assam, India. Biosci Biotech Res Asia 2020;18(1). Available from: https://bit.ly/3sLaINc |
Introduction
The members of class Bivalvia shows unique evolutionary radiation as it displays parental care through brooding of eggs and larva1 and they (Glocidial larva) depend on the species-specific fish host for completion of their life cycle[2]. Taxonomically, the freshwater mussels are placed in the order Unionoida which is the largest group among the five orders of class Bivalvia represented by 6 families, 181 genera and about 840 species. On the other hand, the family Unionidae shows the largest radiation among the 6 families of class Bivalvia that comprises of 142 genera and 620 species worldwide. Though the families of Unionoida are ubiquitously distributed throughout the globe, except Antarctica, this is considered as one of the most threatened groups of freshwater animals alive today 3–5. Being an ecosystem engineer, these freshwater bivalves of order Unionida and family Unionidae play many significant roles to maintain the balance of aquatic ecosystem health[3, 6–14]and need utmost conservation priorities. However, due to the lack of detailed taxonomic and ecological study, application of management and conservation planning is still found to be very difficult. This highlights the urgent need for detailed study in the areas of phylogenetic and evolutionary relationships within the order, family or at the genus level15. Due to their interesting biological characters, for example, reproductive dependence on a host-specific fish for completion of the life cycle, double uniparental inheritance (a particular form of mitochondrial inheritance)16–19 and ecological as well as economic importance, there is a development on phylogenetic studies on freshwater bivalve in recent years [20, 21]. However, conservation efforts focused on species-based and habitat-based unionids are impeded by uncertainties like taxonomic, morphological, ecological, phylogenetic22, 23 and most of the species around the globe have been assigned as Data Deficient (DD) or Least Concerned (LC) species of IUCN Redlist, especially those outside of North America and Western Europe 2, 14, 15, 24 where the majority of phylogenetic study on freshwater unionids have been going on [25, 26]. Recently, several systematic and phylogenetic studies have been carried out on comparatively diverse tropical Asian lineages that have improved our understanding of the classification, morphological evolution, and geographic distribution of many tropical freshwater bivalves 15, 26–28. But still, the south-east Asian and Indo-tropical freshwater unionids have received less attention from the systematic perspective and most of the freshwater unionids taxa remain poorly understood phylogenetically.
India harbours 263 freshwater mollusc species found in two mega biodiversity hotspots, viz. The Western Ghats and Eastern Himalayas of which 95 species are belonging to class Bivalvia14, 29. Out of these total species richness, most of the freshwater bivalves are under DD or LC category of IUCN Redlist. These high numbers of species under DD and LC category are mainly due to unknown species distribution pattern and population trends, lack of information on potential threats and phylogenetic lineages. On the other hand, most of these freshwater mollusc species known only from morphological descriptions of the late 19th and early 20th century. Since then no vast amount of knowledge has been added on the freshwater mollusc species and many morphologically described species seems to be doubtful14, 24, 30, 31. The study on systematics and evolutionary lineages of freshwater unionids in the mainland of India is very limited with the aid of molecular and modern bioinformatics tools32, 33. However, with a single exception of the report by Jadhav and Jamkhedkar 200934 studied the phylogeny of freshwater bivalve, L. corrianus collected from Maharashtra, India, there is absolutely no other report available on DNA sequencing of freshwater mussels based on 18S rRNA sequences from the Assam as well as north-east India. The 18S rRNA is a structural component of the small eukaryotic ribosomal subunit and popularly used in biodiversity research and phylogenetic studies35–38. Compared to conserved cytochrome oxidase subunit I (COI) gene, the 18S rRNA gene is more conserved and its evolutionary progress is much slower making it a suitable molecular marker for phylogenetic studies and for differentiating between taxon at each taxonomic levels 39. Recently, a good number of new species, new information on their biology, bionomics and distribution have been added. A good number of taxonomic account was given by different authors, but at the same time, much confusion was created by the addition of several isolated and inadequate description of species40. So these work needs revision and up to date the present knowledge of freshwater molluscs based phylogenetic studies using sophisticated molecular markers and modern bioinformatics tools. Considering this fact, an attempt was made to a phylogenetic study of two Lamellidens spp. using the amplified sequence of the 18S rRNA gene and to study evolutionary lineage of different Lamellidensspp. concerning available 18S rRNA sequences of other unionids in the global database.
Materials and Methods
Collection and morphological identification Lamellidens specimens
The specimens of the Lamellidens spp. were collected from the upper Brahmaputra basin of Assam covering a total geographical area of approximately 3900 km2 between latitude (27°16’20”–27°47’77”) N and longitude (94°35’30”–95°22’42.16″) E (Figure 1). The large specimens were handpicked and the smaller ones were collected from the bottom substrata by using a metal sieve of mesh size 2mm2. Specimens were then washed, sorted into morpho-species and representatives were brought to the laboratory for future reference. Identification of the recorded specimens was done according to Subba Rao (1989)[41], Ramakrishnan and Dey (2007)[40] and by tallying with authentic voucher specimens deposited at Zoological Survey of India (ZSI), Kolkata.
 |
Figure 1: Map of the study area between latitude 27o16’20”–27o47’77” N and longitude 94o35’30”–95°22’42.16″ E. Coloured dots are different sampling stations of the study area. |
DNA extraction and sequencing of the 18S rRNA gene.
The DNA was extracted from mantle using the Qiagen Blood and Tissue Extraction Kit (Cat No./ ID: 69504). The quality of the extracted DNA from gills and mantle tissue was evaluated in 1% agarose gel. A single band of high molecular weight DNA was observed. Its quantity was determined by measuring the optical density at λ260/230 and λ260/280. After that, the fragment of the 18S rDNA region was amplified by the polymerized chain reaction (PCR) using forward NS1 and reverse NS4 universal primers. A single discrete PCR amplicon band of ~1300 base pairs (bp) was observed when resolved on the agarose gel. The PCR amplicon was purified to remove contaminants. Forward and reverse DNA sequencing reaction of PCR amplicon was carried out with the NS1 and NS4 universal primers using BDT v3.1 Cycle sequencing kit on ABI 3730xl Genetic Analyzer.
Phylogenetic analysis of the sequenced Lamellidens spp.
The consensus sequence was generated from the forward and the reverse sequences, and it was subjected to BLAST-n against non-redundant nucleotide collection. Twenty similar sequences were taken from BLAST result, and the phylogenetic tree was constructed using MEGA 7 software and based on BLAST similarity and phylogenetic affinity of the sequence, the species were identified as L. corrianus and L. phenchooganjensis (Figure 2). The sequences were then deposited to NCBI Genbank and accession numbers MT11611 and MT112199 were obtained for L. corrianus and L. phenchooganjensis respectively.
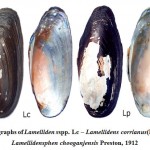 |
Figure 2: Photographs of Lamelliden sspp. Lc – Lamellidens corrianus(Lea, 1834), Lp – Lamellidensphen chooganjensis Preston, 1912. |
Comparison of Lamellidens sequence with other available fresh water unionid bivalves
The partial sequence of the 18S rRNA gene was analysed by Clustal W software to find out the pairwise distance of 18S rRNA gene between Lamellidens spp. and other unionid bivalve sequences present in the nucleotide database of NCBI. A distantly related 18S rRNA sequence was selected as an outgroup to find out the phylogenetic lineages of Lamellidens spp. All the sequences including the outgroup sequence were loaded as FASTA format and multiple sequence alignment (MSA) was performed following Kumar et al. 201642 using MEGA 7 software. After trimming off the flanking ends of the aligned sequences, a phylogenetic tree was constructed using the maximum likelihood method with a bootstrap replication value of 1000.
Result and Discussion
After blast analysis and construction of the phylogenetic tree, it was observed that L. corrianus generated a sequence of about 903 bases and the first 10 hits comprised of partial sequences of 18S rRNA gene of freshwater mussel Lamprotulaleai (Accession No. MF072524), Potomidalittoralis (Accession No. KU763287), Lampsiliscardium (Accession No.KX713305), Elliptiocomplanata (Accession No. JF899209), Aculamprotulatientsinensis (Accession No. MF072525), Thyasira sp. (Accession No. LC187037), Uniopictorum (Accession No. KC429349), Anodontacygnea (Accession No. AM774476), Psiluniolittoralis (Accession No. AF120536) , Elliptiocomplanata (Accession No. AF117738) and Anodonta sp. (Accession No. AY579090). The phylogenetic tree generated showed four major clusters (Figure 3a). The L. corrianus(Accession No. MT111611) forms a cluster with Lamprotulaleai and Potomidalittoralisat the bootstrap confidence level of 77. The three Neotrigoniaspp. (Accession No.s AF 120538, AM774478, and AF411690) forms the second cluster at the bootstrap confidence level of 100. Triplodoncorrugatus (Accession No. KX713352), Hyridellaaustralis (Accession No. KX713301) and Velesunio ambiguous (Accession No. KC429346) formed the third cluster at the bootstrap confidence level of 99. The fourth major cluster was formed by the sequences of freshwater mussels Psiluniolittoralis, Elliptiocomplanata, Aculamprotulatientsinensis, Uniopictorum, Anodontacygneaand Lampsiliscardium.
The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 16 nucleotide sequences and one outgroup sequence of Patella rustica. There were a total of 896 positions in the final dataset.
The L. phenchooganjensis generated a sequence of about 458 bases and after blast analysis and, it was observed that the first 10 hits comprised of partial of 18S rRNA gene of freshwater mussel Aculamprotulatientsinensis, Lamprotulaleai, Lampsilissiliquoidea (Accession No. KY978476), Lampsiliscardium, Fusconaiaflava(Accession No. KX342024), Uniopictorum, Anodontacygnea, Elliptiocomplanata, Psiluniolittoralis etc. The phylogenetic tree generated showed three major clusters (Figure 3b). The three Neotrigoniaspp. forms the first cluster at a bootstrap confidence level of 96. The second cluster comprises Triplodoncorrugatus, Velesunio ambiguous and Hyridellaaustralis at a bootstrap confidence level of 87. The partial 18S rRNA gene sequence of L. phenchooganjensisforms cluster comprises Aculamprotulatientsinensis, Lamprotulaleai, Lampsiliscardium, Uniopictorum, Anodontacygnea, Psiluniolittoralisand Elliptiocomplanata (Figure 3b).
The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 16 nucleotide sequences and one outgroup sequence of Patella rustica. There were a total of 453 positions in the final dataset.
Identification of species is the basis of studies like biological, ecological, genetic, conservation as well as different culture operations. Almost all freshwater mollusc species of Indo-tropical region were identified by the pioneer researchers based on external morphological characters of shells, glocidial larva and internal morphological characters such as the structure of marsupial gills 40, 41 and still, morphological identification has taken as the major tool for the identification of freshwater mussels in India. Although, morphological, anatomical and reproductive keys are found to be important features of basic classification. But, in many instances due to overlapping characters, it is very difficult to make species distinction. Simpson (1900)[43], Modell (1942)44, and Starobogatov (1970)45 had largely used on external shell characters for the establishment of the non-phylogenetic classifications of various new taxa at genus and family level. Since numerous classification and reclassification has been made for unionid bivalve, different and inconsistent classification schemes create many taxonomic confusions and uncertainties in the classification of these freshwater bivalves46.
Some members of genus Lamellidensis more or less widespread in India and some are confined to a limited geographic region within the Indian subcontinent. Due to allopatric distribution pattern and parapatric speciation, the genus display wide range of shell structural variability and colour pattern which remains a challenging task to identify accurately based on morphological, anatomical and reproductive taxonomic keys. Taxonomic keys are still a major problem in establishing a correct database for freshwater mollusc especially the freshwater mussel of India14, 24, 31. With the lake of correct database and inconsistence classification therefore, it is difficult to implement conservation strategies and management measures that result from commercial, municipal and residential development.
From the phylogenetic study of 18S rRNA gene sequencing, it was found that the L. corrianusis closely related to 18S rRNA gene sequences of Lamprotulaleai and Potomidalittoralisat an indent 99.89% and Lampsiliscardium, Elliptiocomplanata, Aculamprotulatientsinensis, Uniopictorum and Anodontacygneaat an indent of around 99.23%. Like L. corrianus, all these species are freshwater mussels belonging to the family Unionidae of order Unionida. After generation of the phylogenetic tree using maximum likelihood method, it was observed that the L. corrianusis phylogenetically neighbour/ very close to Lamprotulaleai and Potomidalittoralis at 77% bootstrap confidence of trees in which the associated taxa are clustered together (Figure 3a). It was also observed that the L. corrianusshowed a high degree of association with the other freshwater mussel from the family Unionidae as branch lengths measured in the number of substitutions per site was very minimum. Therefore, the present study indicates that the L. corrianus showed a single evolutionary lineage with the other considered freshwater mussel species as they showed a common ancestor which was separate from that marine gastropod lineage. The species Patella rustica commonly called a true limpet, a marine gastropod belonging to the family Patellidae was taken as an out group and from the analysis, it was observed that the freshwater mussels and marine gastropods were evolved separately in due course of time from different ancestors as this marine gastropod species group from the rest in the phylogenetic tree (Figure 3a). Similarly, after sequencing analysis of 18S rRNA gene, it was observed that L. phenchooganjensis was also closely related to 18S rRNA gene sequences of Lamprotulaleai, Aculamprotulatientsinensis, Lampsiliscardium, Uniopictorum, Potomidalittoralis and Anodontacygnea with an indent of 99.78% and Elliptiocomplanata and Triplodon corrugates with 99.56% indent. From the analysis of the phylogenetic tree, it was observed that the L. phenchooganjensis was grouped with the above mentioned freshwater mussel species at a bootstrap confidence level of 87% except for Triplodoncorrugatus. The Triplodon corrugates formed a separate group, although the branch length and substitution per site were very less than 0.02 (Figure 3b). Therefore, like other freshwater mussel species, L. phenchooganjensiswas also evolved from a common ancestor which was separate from that of marine lineage as the considered outgroup Patella rusticaformed a separate lineage in the phylogenetic tree (Figure 3b). Though L. corrianusand L. phnechooganjensiswere morphologically distinct species[40, 41], their evolutionary relationship was found to be similar and showed monophyletic origin. Like other freshwater mussels species, the members of Lamellidensspp. shared a common ancestor and showed linear evolutionary lineage (Figure 4).
The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the MCL approach and then selecting the topology with superior log likelihood value. The analysis involved 16 nucleotide sequences and one outgroup sequence of Patella rustica. There were a total of 427 positions in the final dataset.
The result obtained from the phylogenetic analysis of Lamellidens spp. based on 18S rRNA sequencing was found to be significant, because there is only one report of 18S rRNA gene sequence of L. corrianusby Jadhav and Jamkhedkar, 200934 from Maharastra, India. After that, there is no other report about 18S rRNA sequences available/ submitted to the global database except for the present report. On the other hand, 18S rRNA gene sequences of L. phenchooganjensis was submitted for the first time in the Global nucleotide Database (Genebank, NCBI). No previous record on molecular data for this species is available.
According to IUCN, there are 10 globally recognized species under the genus Lamellidens and found to be endemic to Indo-tropical ecoregion. But recent assessment reports suggested that there are only seven species of Lamellidens that are found in mainland of India and some species are still doubtful [40, 47, 48, 49]. For instance, Lamellidensnarain porensisis known only from a single location Kamdai Nadi of Nepal and thought to be possibly a synonym of L. corrianus. Detailed taxonomic study at the molecular level is required to confirm the status of the species47. Similarly, the status as a separate species for Lamellidensunioidesis still doubtful and more taxonomic review is required to confirm it 50. The Lamellidenslamellatusis known only from early 20th century records by Preston (1915)51 and detailed survey and taxonomic study are required to confirm its status as a species, geographical distribution and IUCN Redlist status [48]. The information about the Lamellidensscutumis only based on museum specimens collected only from some localities of Myanmar and India[49]. On the other hand, information on some Lamellidensspp. in global mollusc databases like Molluscabase, Malacolog, UnitasMalacologica is not available or whatever the information available is seem to outdated (Worldwide mollusc species database, Catalogue of Life).
Conclusion
Due to the difficulties in developing a database on population trend, distribution pattern, current geographic range, species coverage and lack of information on potential threats, a comprehensive phylogenetic study on Lamellidens spp. had not been attempted in the past. Therefore the outcome of the present phylogenetic study will help to establish an accurate database and add new information to the existing database especially from those regions where information on freshwater mollusc is considerably lacking.
Acknowledgement
Authors are thankful to Eurofins Scientific, Bengaluru and ZSI, Kolkata for their technical support; DST, Govt. of India for financial support and DST-FIST, Department of Life Sciences for providing necessary facilities for carrying out this work.
References
- Wächtler K., Dreher-Mansur M. C., Richter T.: Larval types and early post larval biology in naiads (Unionoida). In: Ecology and evolution of the freshwater mussels Unionoida. Springer, Berlin, Heidelberg. 2001; pp 93–125.
CrossRef - Bogan A. E., Roe K. J. Freshwater bivalve (Unioniformes) diversity, systematics, and evolution: status and future directions. J. N. Am. Benthol. Soc. 2008; 27: 349-369.
CrossRef - Lydeard C., Cowie R. H., Ponder W. F., Bogan A. E., Bouchet P., Clark S. A., Cummings K. S., Frest T. J., Gargominy O., Herbert D. G., Hershler R., Perez K. E., Roth B., Seddon M., Strong E. E., Thompson F. G. The global decline of nonmarine mollusks. BioSciences 2004; 54: 321-330.
CrossRef - Graf D. L., Cummings K. S. Palaeoheterodont diversity (Mollusca: Trigonoida: Unionoida): What we know and what we wish we knew about freshwater mussel evolution. In: Bivalvia – A Look at the Branches (Bieler R. ed). Zool. J. Linnean. Soc. 2006; 148 pp 343–394.
CrossRef - Bogan A. E. Global diversity of freshwater bivalves (Mollusca:Bivalvia) in freshwater. Hydrobiologia2008; 595: 139-147.
CrossRef - Fenchel T., Kofoed L. H. Evidence for exploitative inter-specific competition in mud snails (Hydrobiidae). Oikos1976; 27: 367-376.
CrossRef - Bertness M. D. 1984 Habitat and community modification by an introduced herbivorous snail. Ecology 1984; 65: 370-381.
CrossRef - Peterson C. H., Black R. Resource depletion by active suspension feeders on tidal fiats: influence of local density and tidal elevation. Limnol. Oceanogr. 1987; 32: 143-166.
CrossRef - Kay E. A. The Conservation Biology of Molluscs. Proceedings of a Symposium Held at the 9th International Malacological Congress, Edinburgh, Scotland, 1995 (No. 9), IUCN.
- Stewart T. W., Miner J. G., Lowe R. L. Quantifying mechanisms for zebra mussel effects on benthic macroinvertebrates: organic matter production and shell–generated habitat. J. N. Am. Benthol. Soc. 1998; 17: 81-94.
CrossRef - Strayer D. L., Caraco N. F., Cole J. J., Findlay S., Pace M. L. Transformation of freshwater ecosystems by bivalves: a case study of zebra mussels in the Hudson River. BioScience 1999; 49: 19-27.
CrossRef - Gutierrez J. L., Jones C. G., Strayer D. L. Iribarne O. O. Mollusks as ecosystem engineers: the role of shell production in aquatic habitats. Oikos 2003; 101: 79-90.
CrossRef - Vaughn C. C., Gido K. B., Spooner D. E. Ecosystem processes performed by unionid mussels in stream mesocosms: species roles and effects of abundance. Hydrobiologia2004; 527: 35-47.
CrossRef - Budha P.B., Aravind N.A., Daniel B.A.: The status and distribution of freshwater molluscs of the eastern Himalaya. In: The Status and Distribution of Freshwater Biodiversity in the Eastern Himalaya, India (Allen DJ, Molur S, Daniel BA, eds). IUCN, 2010; pp 42–53.
- Lopes-Lima M., Froufe E., Ghamizi M., Mock K. E., Kebapçı Ü., Klishko O., Kovitvadhi S., Kovitvadhi U., Paulo O. S., Pfeiffer III J. M., Raley M. Phylogeny of the most species–rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): Defining modern subfamilies and tribes. Mol. Phylogenetics Evol. 2017; 106: 174-191.
CrossRef - Hoeh W. R., Black M. B., Gustafson R., Bogan A. E., Lutz R., Vrijenhoek R. C. Testing alternative hypotheses of Neotrigonia (Bivalvia: Trigonioida) phylogenetic relationships using cytochrome c oxidase subunit I DNA sequences. Malacologia 1998; 40: 267–278.
- Hoeh W. R., Stewart D. T., Guttman S. I. High fidelity of mitochondrial genome transmission under the doubly uniparental mode of inheritance in freshwater mussels (Bivalvia: Unionoidea). Evolution 2002; 56: 2252-2261.
CrossRef - Barnhart M.C., Haag W.R., Roston W.N. Adaptations to host infection and larval parasitism in Unionoida. J. N. Am. Benthol. Soc. 2008; 27: 370–394.
CrossRef - Breton S., Beaupre H.D., Stewart D.T., Hoeh W.R., Blier P.U. The unusual system of doubly uniparental inheritance of mtDNA: isn’t one enough?. Trends Genet. 2007; 23: 465–474.
CrossRef - Haag W.R. (ed): North American freshwater mussels: natural history, ecology, and conservation. Cambridge University Press. 2012
CrossRef - Lopes-Lima M., Teixeira A., Froufe E., Lopes A., Varandas S., Sousa R. Biology and conservation of freshwater bivalves: past, present and future perspectives. Hydrobiologia 2014; 735: 1–13.
CrossRef - Inoue K., McQueen A.L., Harris J.L., Berg D.J. Molecular phylogenetics and morphological variation reveal recent speciation in freshwater mussels of the genera Arcidens and Arkansia (Bivalvia: Unionidae). Biol. J. Linn. Soc. 2014; 112, 535–545.
CrossRef - Pfeiffer J.M., Graf D.L. Evolution of bilaterally asymmetrical larvae in freshwater mussels (Bivalvia: Unionoida: Unionidae). Zool. J. Linnean Soc.2015; 175: 307–318.
CrossRef - Köhler F., Seddon M., Bogan A.E., Tu D.V., Aroon P.S., Allen D. The status and distribution of freshwater molluscs of the Indo-Burma region. In: The Status and Distribution of Freshwater Biodiversity in Indo–Burma (Allen DJ, Smith KG, Darwall WRT, eds.). IUCN: Cambridge, UK and Gland, Switzerland; Zoo Outreach Organization, Coimbatore, India. 2012; pp 67–85.
- Lopes-Lima M., Burlakova, L.E., Karatayev, A.Y., Mehler, K., Seddon, M. & Sousa, R. (2018) Conservation of freshwater bivalves at the global scale: diversity, threats and research needs. Hydrobiologia 810, 1–14.
CrossRef - Bolotov I.N., Pfeiffer J.M., Konopleva E.S., Vikhrev I.V., Kondakov A.V., Aksenova O.V., Gofarov M.Y., Tumpeesuwan S., Win T. A new genus and tribe of freshwater mussel (Unionidae) from Southeast Asia. Scientific reports 2018; 8: 1–12.
CrossRef - Zieritz A., Bogan A.E., Froufe E., Klishko O., Kondo T., Kovitvadhi U., Kovitvadhi S., Lee J.H., Lopes–Lima M., Pfeiffer J.M., Sousa R. Diversity, biogeography and conservation of freshwater mussels (Bivalvia: Unionida) in East and Southeast Asia. Hydrobiologia 2018; 810: 29–44.
CrossRef - Konopleva E.S., Bolotov I.N., Vikhrev I.V., Gofarov M.Y., Kondakov A.V. An integrative approach underscores the taxonomic status of Lamellidensexolescens, a freshwater mussel from the Oriental tropics (Bivalvia: Unionidae). Systematics and Biodiversity 2017; 15: 204–217.
CrossRef - Aravind N.A., Madhyastha N.A., Rajendra G.M., Dey A. The status and distribution of freshwater molluscs of the Western Ghats. In: The Status and Distribution of Freshwater Biodiversity in the Western Ghats, India (Allen DJ, Molur S, Daniel BA, eds). IUCN, 2011; pp 49–62.
- Kumar A., Vyas V. Diversity of Molluscan community in River Narmada, India. J. Chem. Biol. Phys. Sci. 2012; 2: 1407–1412.
- Ramesha M.M., Sophia S., Muralidhar, M. Freshwater bivalve fauna in the Western Ghats Rivers of Karnataka, India: Diversity, distribution patterns, threats and conservation needs. Int. J. Cur. Res. 2013; 5: 2500–2505.
- Upadhye M.V. Patil R.C. Manohar S.M., Jadhav U. Phylogenetic Study of Freshwater Bivalve ParreysiaCorrugata from Maharashtra State, India by 18S rRNA Sequences. J. Life Sci. 2011; 5: 733–738.
- Magare V.N., Kulkarni C.P., Maurya C.B., Patil R.C., Upadhye M.V. Phylogenetic analysis of freshwater mussel Corbicula regularisby 18S rRNA gene sequencing. J. Exp. Biol. Agri. Sci. 2015; 3: 213–219.
- Jadhav B.L., JamkhedkarS. Phylogenetic Analysis of Lamellidenscorrianus obtained from Konkan Region of Maharashtra by 28s rRNA and 18s rRNA sequences. Res. J. Biotech. 2009; 4, 37–44.
- Šlapeta J., Moreira D., López-García P. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc. R. Soc. B: Biol. Sci. 2005; 272: 2073–2081.
CrossRef - Krüger M., Krüger C., Walker C., Stockinger H., Schüßler A. Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol 2012; 193: 970–984.
CrossRef - Buse H.Y., Lu J., Struewing I.T., Ashbolt N.J. Eukaryotic diversity in premise drinking water using 18S rDNA sequencing: implications for health risks. Environ. Sci. Pollut. Res. 2013; 20: 6351–6366.
CrossRef - Fonseca V.G., Carvalho G.R., Nichols B., Quince C., Johnson H.F., Neill S.P., Lambshead J.D., Thomas W.K., Power D.M., Creer S. Metagenetic analysis of patterns of distribution and diversity of marine meiobenthic eukaryotes. Glob. Ecol. Biogeogr. 2014; 23: 1293–1302.
CrossRef - Tang C.Q., Leasi F., Obertegger U., Kieneke A., Barraclough T.G., Fontaneto D. The widely used small subunit 18S rDNA molecule greatly underestimates true diversity in biodiversity surveys of the meiofauna. Proc. Natl. Acad. Sci. 2012; 109: 16208–16212.
CrossRef - Ramakrishna, Dey A.: Handbook on Indian Freshwater Molluscs. Zoological Survey of India, Kolkata. 2007.
- Subba Rao N.V.: Handbook on Indian Freshwater Molluscs. Zoological Survey of India, Kolkata. 1989
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016; 33(7): 1870-1874.
CrossRef - Simpson C.T. Synopsis of the naiades, or pearly freshwater mussels. Proc. U. S. Natl. Mus. 1900; 22: 501–1044.
CrossRef - Modell H. Das natürliche system der najaden. Arch. Molluskenkd. 1942; 74: 161–191.
- Starobogatov Y.I.: Molluscan Fauna and Zoogeographic Zonation of Continental Freshwater Bodies of the World. Leningrad: ZoologiceskijInstitut, AkademijaNauk SSSR, 1970.
- Hoeh W.R., Bogan A.E., Heard W.H.: A phylogenetic perspective on the evolution of morphological and reproductive characteristics in the Unionoida. In: Ecology and evolution of the freshwater mussels Unionoida. Springer, Berlin, Heidelberg. 2001; pp 257–280.
CrossRef - Budha P.: Lamellidensnarainpirensis. The IUCN Red List of Threatened Species 2010, e.T173030A6961298. https://dx.doi.org/ 10.2305/ IUCN.UK.2010–4.RLTS.T173030A6961298.en.
CrossRef - Budha P.B., Daniel B.A.: Lamellidenslamellatus. The IUCN Red List of Threatened Species 2010, e.T166687A6259747. http:// dx.doi.org/ 10.2305/IUCN.UK.2010–4.RLTS.T166687A6259747.en
CrossRef - Madhyastha A., Daniel B.A., Bogan, A.E.: Lamellidensscutum. The IUCN Red List of Threatened Species 2014, e.T173032A1376335. https://dx.doi.org/10.2305/IUCN.UK.2014–3.RLTS.T173032A1376335.en.
CrossRef - Madhyastha A.: Lamellidensunioides. The IUCN Red List of Threatened Species 2010, e.T177460A7439640. https://dx.doi.org/ 10.2305/IUCN.UK.2010–4.RLTS.T177460A7439640.en.
CrossRef - Preston H.B.: The Fauna of British India including Ceylon and Burma. Mollusca (Freshwater Gastropoda and Pelecypoda) Taylor and Francis, London, 1915.

This work is licensed under a Creative Commons Attribution 4.0 International License.








