How to Cite | Publication History | PlumX Article Matrix
Om Kumar , Keerthana G<
, Keerthana G< , Ashitha B Arun
, Ashitha B Arun , Ananya Joliholi
, Ananya Joliholi  and Lokesh Ravi*
and Lokesh Ravi*
Department of Botany, St. Joseph’s College (Autonomous), Bengaluru, India.
Corresponding Author E-mail: lokesh.ravi@sjc.ac.in
DOI : http://dx.doi.org/10.13005/bbra/2943
ABSTRACT:
The aim of this study is to construct 3D models of potential drug targets for the Bacterial Kidney Disease (BKD) causing pathogen Renibacterium salmoninarum. The bacterial pathogen Renibacterium salmoninarum was selected for homology modeling studies since there were no known protein structures of the organism reported in the NCBI database. The reported protein sequences were run through DrugBank to pick out drug-targets. Online databases and web tools such as PMDB, UniProt, Drug Bank, and SwissModel were employed in this analysis. An aggregate of 412 protein sequences were identified as potential drug targets and were retrieved from the UniProt. Homology models of the protein sequences were constructed using the SwissModel database for all 412 proteins. These were then refined through a protein blast and Ramachandran plot analysis. Out of the 412 constructed models, 143 models were of reliable quality. These were then submitted to the PMDB database for further reference. To demonstrate the application of these constructed models, protein-ligand docking analysis using Auto Dock Vina was performed. Among the antibiotics that were tested against their known drug targets, trimethoprim demonstrated significant potential for the inhibition of R. salmoninarum’s dihydrofolate reductase protein, with a binding energy of -9.06 Kcal/mol and with the formation of 3 hydrogen bonds. Therefore through protein-ligand docking studies and the construction of 3D models of protein drug targets, Trimethoprim is proposed as a solution to the Bacterial Kidney Disease (BKD) problem in salmonid fishes. Further in-vitro evidences are in demand to prove this hypothesis.
KEYWORDS: Bacterial Kidney Disease; Homology Modelling; Protein Drug Targets; Protein Model Database; Renibacterium salmoninarum
Download this article as:| Copy the following to cite this article: Kumar O, Keerthana G, Arun A. B, Joliholi A, Ravi L. Construction of 3D Model of Protein Drug Targets for Renibacterium Salmoninarum - a Bacterial Pathogen Causing Bacterial Kidney Disease in Young Salmonid Fish. Biosci Biotech Res Asia 2021;18(3). |
| Copy the following to cite this URL: Kumar O, Keerthana G, Arun A. B, Joliholi A, Ravi L. Construction of 3D Model of Protein Drug Targets for Renibacterium Salmoninarum - a Bacterial Pathogen Causing Bacterial Kidney Disease in Young Salmonid Fish. Biosci Biotech Res Asia 2021;18(3). Available from: https://bit.ly/3pJWKfH |
Introduction
Salmon is the common name used to refer to the various species of the Salmonidae family. There are nine commercially important species of salmon and these are widespread across the world. Most are found in the Pacific and Atlantic oceans, contributing significantly to the economic and cultural value of the nations situated near these oceans. For example, Rawas, also known as the Indian salmon, the only species that can be found in India, (and primarily spotted in the waters of the Indian states of Gujarat and Maharashtra) is a huge contributor to the income generated here. In 2010, the harvesting, processing, and retailing of the Bristol Bay Salmon (Oncorhynchus kisutch) created $1.5 billion in sales value across the United States.1 Norway is thought to be the largest contributor to the Atlantic Salmon Industry. The country accounted for a total of 63% (2.3 million tonnes) of the total production of Salmon in 2016.
Farmed salmon is now a global commodity which has seen an exponential rise in various markets.2 Salmon aquaculture has expanded rapidly within the last two decades. It is predicted that salmon aquaculture will continue to expand in order to meet growing seafood demand, since wild capture fisheries have stagnated in terms of production. In Norway, this is a government backed industry with high levels of innovation.3 The production of the farmed species of salmon is restricted to a few countries like Norway, Chile, the UK and Canada. Nevertheless, this contributes a whopping 85% to worldwide production. Salmon aquaculture is responsible for 15% of the United Kingdom’s agricultural economy.
Salmon fishes are a favourite with culinary chefs, and are liked for their orange-coloured meat. The meat is believed to have several health benefits, including reducing obesity, making it extremely valuable as seafood.
Renibacterium salmoninarum is responsible for causing disease in young Salmonid fish. The infection is commonly known as Bacterial Kidney Disease (BKD), Dee Disease, White Boil Disease or the Corynebacterial Kidney Disease. It is of substantial ecological importance due to its effect on both farmed and wild Salmonids. It was first described under the name of Dee disease (1930), and was identified in the Atlantic salmon (Salmo salar) species in Scotland. A gram positive diplobacillus bacterium that did not grow on any available media was recognized in the kidneys of the diseased fish. This pathogen was found to be exclusive to Salmonids. Ordal and Earp initially cultured the bacteria and identified it as a species of Corynebacterium based on its morphological appearance. Smith concluded that the Dee disease of salmonids in Scotland and the Bacterial Kidney Disease were caused by the same bacterium. 4, 5
Government agencies for statutory fish health programmes in the UK (FRS Marine Laboratory, Aberdeen and Cefas, Weymouth) participated in a comparative study for the detection of Renibacterium salmoninarum. FRS conducted additional tests – Indirect fluorescent antibody (IFAT), Gram staining of tissue sections and H&E, a quantitative real-time PCR (qPCR) assay, to detect the elongation factor alpha 1 (ELF) gene of salmonids as well as the Msa2 genes of R. salmoninarum. Isolation using Mueller Hinton with added cysteine (MHCA) resulted in the fish testing to be culture positive. 6
Renibacterium salmoninarum is an intracellular pathogen which multiplies within phagocytes. It is slow growing and is described as a coherent genus. The properties associated with the cell envelope demonstrate the intracellular survival and multiplication characteristics in phagocytic cells.
The cell surface hydrophobicity of R. salmoninarum strains, examined using a salt aggregation method showed the strains to be sticky, auto-agglutinating, and were found to possess a hydrophobic cell surface. Strains with low virulence were found to be non-agglutinating and non-sticky. The adherence of the bacteria to host tissues plays a significant role in their ability to colonize and cause infection.7
Renibacterium salmoninarum is a sessile, strongly Gram-positive, non acid fast, non spore forming, rod shaped bacterium that usually occurs in pairs (diplococcus). It is approximately 0.3-1.0 µm × 1.0-1.5 µm in length.8 A slow growing organism, it is one of the earliest known bacterial pathogens of fish (especially salmonids). It is a facultative intracellular parasite, and because of this intracellular nature of infection, the bacterium has the ability to evade the immune response of the host. This makes BKD an especially challenging disease to control.9
The cell wall of R. salmoninarum is composed primarily of peptidoglycan. The major sugar component is galactose, in addition to N-acetylfucosamine, rhamnose and N-acetylglucosamine. Major amino acids present are alanine, glutamic acid, lysine, and glycine, where the third position of the peptide subunit of the peptidoglycan contains lysine. There is an interpeptide bridge between the lysine and the D-alanine of adjacent peptide subunits that is composed of glycylalanine.
R. salmoninarum is very similar to the Coryneform group of bacteria.8,10 The bacterium lives inside the pronephric kidneys of salmonid fish. It spreads in two ways – vertically inside the ova, and horizontally in shared water between cohabiting fish. A general consensus is that R. salmoninarum has co-evolved along with its host, the salmonids.10,11 Diagnostic tools for BKD are PCRs, especially real time quantitative PCRs (RT qPCRs).11
Control of infection of R. salmoninarum is an arduous process since there are no vaccines available that are sufficiently effective. Antibiotic resistance is also a significant complication with this pathogen. 9,12
In spite of being a potent pathogen causing significant economic damage to the fish industry, specifically to salmon and trout8, very little information is available about this bacterial pathogen. It has been studied very little mainly due to the punctilious and slow growing nature of the disease. This makes researching R. salmoninarum difficult. A more thorough study of R. salmoninarum remains necessary to combat the evasive Bacterial Kidney Disease.
Homology Modelling is a computational technique that predicts the structure of a protein sequence by comparing it to the structure of a homologous protein sequence, on the basis of degree of similarity. In this study, the 3D structures of the proteins of Renibacterium salmoninarum were developed using the technique of homology modelling. The protein structures are essential in order to design and develop drugs that can be used to curb the spread of this disease. The lack of availability of protein structures has hindered the understanding of binding specificities of proteins and ligands, which are prerequisites for drug design and development13,14. Well Established and recognized databases and tools were utilized for computing structures of potential drug target proteins. This data is important to curb the spread of this disease and also reduce the financial burdens incurred on the fisheries and the pisciculture industry on account of this disease. Homology modelling was carried out using preexisting FASTA format sequences of the proteins involved, along with similar preexisting templates that closely resembled the query sequence, in order to predict the structure of the proteins. It is absolutely necessary for the protein sequences to be available in order to understand the binding specificities of the protein and ligand to yield higher amounts of drugs to curb the spread of the disease. The structures developed in this study can be further exploited to develop drugs, which will be a boon to the fisheries involved in the rearing of salmonoid fishes13.
Materials and Methods
NCBI Database
The pre-existing information available on the organism was extracted from the NCBI database. The database compiles data about the organism from multiple databases (https://www.ncbi.nlm.nih.gov/).
Sequence Retrieval
The amino acid sequences required for this study were collected from the Uniprot Knowledge Database. The database is centralized, reliable and publicly accessible. The sequences with the best match were sorted based on the length of the amino acid chain. The required amino acid chain files were downloaded in the FASTA file format. The downloaded sequences were then saved with their respective accession IDs. This was for enabling easier accessibility in the future. The database can be accessed at: www.uniprot.org.
Sequence Alignment
The UNIPROT database enabled the compilation of amino acid sequences. The sequences were compared using the BLASTp server. This permits the comparison of the query sequence (also known as the amino acid sequence) with pre-existing protein sequences in the Protein Data Bank (for gauging percentage similarity). The first BLAST was performed for getting values similar to that of the pre-existing sequences of the non-redundant database. This was followed by a second BLAST which was conducted to obtain similar values with the sequences present in the Protein Data Bank (www.rcsb.org). The percentage of similarity was documented for further reference.
Structure Prediction
The Homology modelling technique was performed using the SWISS MODEL tool (https://swissmodel.expasy.org/). This online tool helped predict the three-dimensional structures of the selected drug-target proteins. The respective amino acid sequences of the proteins, as well as the templates available in the protein data bank were used to predict the 3D structure of the protein. The sequences obtained from UNIPROT were uploaded as FASTA files and the output (3D structures) were stored in PDB format. The availability and percentage similarity of the templates were the important factors that influenced the developed models. Ramachandran plots were used to analyze the structures. These are graphical plots that help in confirming the predicted structure’s accuracy. The most accurate of models were downloaded in PDB format.
Model Analysis
Qualitative analysis of the accuracy of the predicted model was conducted using the built-in Ramachandran plot feature in SWISSMODEL. It was expected that the degree angles of all the residues would be found within the Most Favored regions of the Ramachandran plot, as this is what determines the quality of the predicted structure. The residues that were observed outside of this favored region were considered to be unfavorable. These outliers affected the confidence score of the predicted model.
Model Submission
The 3D models of the proteins that exhibited good quality Ramachandran plot (confidence score) were submitted to the Protein Model Database (PMDB) (http://srv00.recas.ba.infn.it/PMDB/). The Protein Model Database is a resource that harbors manually built protein models that have been published in research based journals. All the models were uploaded in pdb file format and each entry was given a unique PMDB ID for future reference.
Results and Discussion
Pathogen and Drug Target Selection
The pathogen Renibacterium salmoninarum was identified as a potential target for homology modeling studies since there were no known protein structures of the organism present in the NCBI database (although there were 1309 reported protein sequences available). Thus, R. salmoninarum was ideal for computational protein model creation. Among the 1309 protein sequences, non-enzymatic proteins, subunits as well as duplicates were eliminated, leaving a final count of 1249 proteins. These 1249 proteins were then verified on the DrugBank website (www.drugbank.ca) in order to check if these were potential drug targets. Among the 1249 proteins, a total of 412 protein sequences were identified as drug targets. Homology model construction was carried out for these 412 protein sequences. Many similar studies have seen a rise due to the coronavirus outbreak, homology modelling and molecular docking have been key in identifying drug targets and potential medicines. A homology modelling study of TMPRSS2( spike entry priming) was done using a similar pathway.
Constructing Homology Models
The 412 selected protein sequences were retrieved from the UniProt database (www.uniprot.org) and were stored in the FASTA file format (. fasta). These sequences were then subjected to a BLASTp (also called as Protein Blast) (https://www.blast.ncbi.nlm.nih.gov/) to find within the PDB website (www.rcsb.org), suitable templates for homology modelling for the protein structures that had more than 80% similarity match. All of the 412 sequences showed a match of greater than 80% with the available crystal structures in the PDB website. Therefore, all the 412 sequences were subjected to computational homology modelling using SWISS Model (https://swissmodel.expasy.org/). The tool created different models for each protein sequence. Among the created protein sequences modelled, the optimal model was selected using the Ramachandran plot analysis.
Ramachandran Plot Analysis
Ramachandran plot analysis was carried out on all the protein sequence models constructed using SWISSModel, via inbuilt options in the website. The protein models which portrayed more than 95% of residues in the favoured regions of the Ramachandran plot were considered to be eligible for structural application. Among the 412 constructed proteins, 143 protein models contained more than 95% of residues in the Ramachandran favoured region and were thus considered for further applications. Fig.1 shows the graphical representation of the most preferred protein model with 100% score in the favoured region and a least preferred model with 80% score within the favoured region.
PMBD ID Submission
Constructed protein models of all the 143 proteins were then submitted to a public database (PMDB) (http://srv00.recas.ba.infn.it/PMDB/) for public access. A catalogue of the constructed protein models that were submitted to the PMDB database is listed in Table.1. This allows researchers working in this field of study to retrieve and execute further structural bioinformatics analyses specific to this pathogen.
Table 1: PMDB entries for Renibacterium salmoninarum. The following drug targets and their respective UniProt Ids, the Ramachandran favoured region and the PMDB IDs have been used in this study to determine the potential drug targets for R. salmoninarum. The Ramachandran scores above 95% were considered to obtain the potential structure of proteins.
| Sl. No. | Drug Target Name | UniProt ID | Ramachandran Favourable Region | PMDB ID |
| 1 | DHNA-CoA synthase | A9WLY2 | 95.96% | PM0083536 |
| 2 | DXP reductoisomerase | A9WML8 | 95.49% | PM0083539 |
| 3 | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | A9WTJ3 | 96.76% | PM0083543 |
| 4 | BPG-dependent PGAM | A9WSB8 | 97.00% | PM0083530 |
| 5 | 2,5-diketo-D-gluconic acid reductase | A9WQX7 | 95.27% | PM0083548 |
| 6 | MECDP-synthase | A9WLY3 | 96.54% | PM0083556 |
| 7 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | A9WLY2 | 96.32% | PM0083557 |
| 8 | 2-isopropylmalate synthase | A9WML8 | 95.80% | PM0083559 |
| 9 | 3-dehydroquinase | A9WTJ3 | 97.65% | PM0083560 |
| 10 | 3-dehydroquinate synthase | A9WSB8 | 95.34% | PM0083564 |
| 11 | 3-hydroxyisobutyryl-CoA hydrolase | A9WQX7 | 96.10% | PM0083565 |
| 12 | Ketopantoate hydroxymethyltransferase | A9WS61 | 95.38% | PM0083567 |
| 13 | HMBPP reductase | A9WPD7 | 96.38% | PM0083586 |
| 14 | HTPA reductase | A9WQ18 | 96.28% | PM0083591 |
| 15 | HTPA synthase | A9WNM3 | 95.76% | PM0083593 |
| 16 | DMRL synthase | A9WR65 | 96.34% | PM0083596 |
| 17 | 6-phosphogluconate dehydrogenase | A9WV17 | 95.97% | PM0083597 |
| 18 | Acetyl-CoA acetyltransferase | A9WSQ3 | 95.17% | PM0083598 |
| 19 | Adenosylhomocysteinase | A9WU61 | 96.19% | PM0083599 |
| 20 | Adenylate kinase | A9WST7 | 97.86% | PM0083600 |
| 21 | Adenylosuccinate synthetase | A9WV79 | 96.69% | PM0083605 |
| 22 | Aldo/keto reductase | A9WQQ7 | 96.21% | PM0083606 |
| 23 | Alpha-methylacyl-CoA racemase | A9WST4 | 96.23% | PM0083608 |
| 24 | Amino acid transporter | A9WNN1 | 95.42% | PM0083610 |
| 25 | Ammonium transporter | A9WP34 | 95.55% | PM0083612 |
| 26 | Anthranilate phosphoribosyltransferase | A9WR05 | 96.02% | PM0083614 |
| 27 | Argininosuccinate synthase | A9WQ90 | 96.82% | PM0083616 |
| 28 | Aspartate-semialdehyde dehydrogenase | A9WNH0 | 95.58% | PM0083618 |
| 29 | Aspartate–tRNA(Asp/Asn) ligase | A9WSD4 | 97.22% | PM0083620 |
| 30 | ATP phosphoribosyltransferase | A9WR63 | 98.56% | PM0083622 |
| 31 | ATP-dependent dethiobiotin synthetase | A9WL42 | 96.13% | PM0083623 |
| 32 | ATP-dependent RNA helicase | A9WPJ4 | 95.90% | PM0083625 |
| 33 | ATP-dependent RNA helicase | A9WNH6 | 96.08% | PM0083629 |
| 34 | Bacterial regulatory protein | A9WTC5 | 96.49% | PM0083631 |
| 35 | Beta-thionase | A9WQJ7 | 95.36% | PM0083632 |
| 36 | Bifunctional protein | A9WU39 | 96.16% | PM0083633 |
| 37 | Bifunctional purine biosynthesis protein | A9WLT2 | 96.24% | PM0083635 |
| 38 | Branched-chain amino acid aminotransferase | A9WNW7 | 95.43% | PM0083637 |
| 39 | Carbonic anhydrase | A9WME9 | 96.56% | PM0083642 |
| 40 | Chaperone protein | A9WQP7 | 100.00% | PM0083705 |
| 41 | Chitosanase | A9WUI6 | 96.17% | PM0083708 |
| 42 | Chorismate mutase | A9WMQ5 | 100.00% | PM0083966 |
| 43 | Chorismate synthase | A9WSC0 | 96.93% | PM0083710 |
| 44 | Citrate synthase | A9WQB8 | 96.17% | PM0083890 |
| 45 | Cystathionine gamma-lyase | A9WQJ8 | 96.05% | PM0083891 |
| 46 | Cytidylate kinase | A9WRN3 | 97.27% | PM0083892 |
| 47 | dCTP deaminase | A9WT08 | 97.33% | PM0083893 |
| 48 | Delta-aminolevulinic acid dehydratase | A9WPH8 | 95.65% | PM0083955 |
| 49 | Deoxyribodipyrimidine photolyase | A9WN66 | 97.86% | PM0083934 |
| 50 | Deoxyribose-phosphate aldolase | A9WU17 | 97.02% | PM0083935 |
| 51 | Diaminopimelate decarboxylase | A9WNA8 | 95.31% | PM0083936 |
| 52 | Dihydrofolate reductase | A9WNH1 | 96.45% | PM0083938 |
| 53 | Dihydroorotate dehydrogenase | A9WML3 | 95.38% | PM0083939 |
| 54 | Dihydropteroate synthase | A9WUQ9 | 97.19% | PM0083940 |
| 55 | DNA ligase | A9WTZ3 | 97.53% | PM0083941 |
| 56 | DNA-3-methyladenine glycosylase | A9WPF0 | 97.71% | PM0083942 |
| 57 | Enoyl-CoA hydratase | A9WVC8 | 96.19% | PM0083943 |
| 58 | Farnesyl pyrophosphate synthetase | A9WRS8 | 95.31% | PM0083944 |
| 59 | Fibrillar surface protein | A9WSZ8 | 95.33% | PM0083945 |
| 60 | Fumarylacetoacetate | A9WLG1 | 95.00% | PM0083946 |
| 61 | Gamma-glutamyltranspeptidase | A9WL74 | 99.38% | PM0083947 |
| 62 | GDP-mannose 4,6 dehydratase | A9WN78 | 95.16% | PM0083948 |
| 63 | Glucan endo-1,3-beta-glucosidase | A9WSK0 | 98.51% | PM0083949 |
| 64 | Glucosamine-6-phosphate deaminase | A9WNE3 | 96.90% | PM0083950 |
| 65 | Glucose-6-phosphate 1-dehydrogenase | A9WT42 | 95.49% | PM0083951 |
| 66 | Glutamate racemase | A9WNP4 | 97.28% | PM0083952 |
| 67 | Glutamate-1-semialdehyde aminotransferase | A9WPI0 | 96.02% | PM0083953 |
| 68 | Glutamyl-tRNA synthetase | A9WP26 | 97.11% | PM0083954 |
| 69 | D-fructose-6-phosphate amidotransferase | A9WMF4 | 95.19% | PM0083956 |
| 70 | Glutaryl-CoA dehydrogenase (EC 1.3.8.6) | A9WQK3 | 96.55% | PM0083957 |
| 71 | Glycerol kinase | A9WS93 | 96.39% | PM0083958 |
| 72 | Glycerol-3-phosphate dehydrogenase | A9WMG6 | 95.28% | PM0083959 |
| 73 | Glycine dehydrogenase | A9WME4 | 95.47% | PM0083960 |
| 74 | Glutamine amidotransferase | A9WN01 | 95.46% | PM0083961 |
| 75 | GTP cyclohydrolase 1 | A9WUR0 | 96.47% | PM0083962 |
| 76 | Guanylate kinase | A9WSA4 | 97.77% | PM0083963 |
| 77 | Guanyl-specific ribonuclease | A9WLY7 | 100.00% | PM0083964 |
| 78 | Haloacid dehalogenase-like hydrolase | A9WV91 | 95.93% | PM0083965 |
| 79 | Histidine ammonia-lyase | A9WN95 | 96.09% | PM0083652 |
| 80 | ATP-dependent DNA helicase | A9WSE4 | 97.76% | PM0083578 |
| 81 | Homoserine O-acetyltransferase | A9WMJ0 | 95.67% | PM0083581 |
| 82 | Hypoxanthine phosphoribosyltransferase | A9WUR5 | 96.87% | PM0083583 |
| 83 | Lipoyl synthase | A9WS40 | 97.67% | PM0083585 |
| 84 | L-serine dehydratase | A9WME8 | 95.68% | PM0083587 |
| 85 | Lysine-tRNA ligase | A9WUP4 | 95.84% | PM0083590 |
| 86 | Malate synthase | A9WLG6 | 96.88% | PM0083595 |
| 87 | Malonyl-CoA-[acyl-carrier-protein] transacylase | A9WP65 | 95.36% | PM0083603 |
| 88 | Methionine aminopeptidase | A9WM52 | 95.69% | PM0083607 |
| 89 | Methionine–tRNA ligase | A9WP14 | 98.03% | PM0083617 |
| 90 | Methylisocitrate lyase | A9WQB9 | 96.63% | PM0083626 |
| 91 | Molybdopterin biosynthesis enzyme | A9WQ16 | 96.05% | PM0083653 |
| 92 | NH(3)-dependent NAD(+) synthetase | A9WUC0 | 98.70% | PM0083654 |
| 93 | Non-specific serine/threonine protein kinase | A9WTU4 | 97.76% | PM0083655 |
| 94 | Nucleoside diphosphate kinase | A9WUV0 | 97.24% | PM0083656 |
| 95 | Organic hydroperoxide resistance protein | A9WMD8 | 95.54% | PM0083657 |
| 96 | Orotate phosphoribosyltransferase | A9WV95 | 96.33% | PM0083658 |
| 97 | o-succinylbenzoate synthase | A9WRT7 | 95.71% | PM0083659 |
| 98 | Pantothenate kinase | A9WMF5 | 96.41% | PM0083660 |
| 99 | Phospho-2-dehydro-3-deoxyheptonate aldolase | A9WRF8 | 96.00% | PM0083661 |
| 100 | Phosphoenolpyruvate carboxykinase | A9WL73 | 95.17% | PM0083662 |
| 101 | Phosphoenolpyruvate-protein phosphotransferase | A9WUJ2 | 98.81% | PM0083663 |
| 102 | Phosphoglycerate mutase family protein | A9WVN2 | 95.08% | PM0083664 |
| 103 | Phosphopantetheine adenylyltransferase | A9WMZ3 | 96.08% | PM0083665 |
| 104 | Phosphoribosylformylglycinamidine cyclo-ligase | A9WQZ0 | 96.24% | PM0083666 |
| 105 | Phosphoribosylglycinamide formyltransferase | A9WLR9 | 95.97% | PM0083667 |
| 106 | Predicted secreted protein | A9WPE9 | 100.00% | PM0083668 |
| 107 | Probable nicotinate-nucleotide adenylyltransferase | A9WUT8 | 96.83% | PM0083669 |
| 108 | Recombinase A | A9WQ43 | 95.40% | PM0083969 |
| 109 | Purine nucleoside phosphorylase | A9WU15 | 95.91% | PM0083970 |
| 110 | Putative quinone oxidoreductase | A9WLL0 | 95.49% | PM0083971 |
| 111 | Pyridoxamine 5′-phosphate oxidase | A9WQ92 | 95.89% | PM0083974 |
| 112 | Pyruvate kinase | A9WR48 | 95.51% | PM0083973 |
| 113 | Quinone oxidoreductase | A9WQ80 | 96.91% | PM0083975 |
| 114 | Regulatory protein | A9WQ45 | 100.00% | PM0083976 |
| 115 | Ribokinase | A9WUQ1 | 95.65% | PM0083977 |
| 116 | Ribose 5-phosphate isomerase | A9WUW8 | 97.70% | PM0083978 |
| 117 | Ribulose-phosphate 3-epimerase | A9WR72 | 95.39% | PM0083979 |
| 118 | S-adenosylmethionine synthase | A9WSA1 | 96.28% | PM0083980 |
| 119 | Serine hydroxymethyltransferase | A9WM39 | 96.39% | PM0083981 |
| 120 | Serine–tRNA ligase | A9WUS1 | 97.40% | PM0083982 |
| 122 | Signal recognition particle receptor | A9WP32 | 95.72% | PM0083983 |
| 123 | Sodium/proline transporter | A9WL16 | 96.94% | PM0083984 |
| 124 | Spermidine synthase | A9WUF9 | 96.15% | PM0083985 |
| 125 | SURF1-like protein | A9WSP5 | 100.00% | PM0083986 |
| 126 | Thioredoxin peroxidase | A9WSN6 | 98.65% | PM0083987 |
| 127 | Thiosulfate sulfurtransferase | A9WPZ8 | 96.15% | PM0083988 |
| 128 | Thymidine phosphorylase | A9WKS1 | 97.85% | PM0083989 |
| 129 | Thymidylate synthase | A9WNH2 | 96.12% | PM0084073 |
| 130 | Transaldolase | A9WT40 | 97.24% | PM0083990 |
| 131 | Transcriptional regulator | A9WT73 | 96.32% | PM0084022 |
| 132 | Transcriptional regulator | A9WMH1 | 95.85% | PM0084021 |
| 133 | Tryptophan 2,3-dioxygenase | A9WT09 | 98.70% | PM0084023 |
| 134 | Tryptophan synthase alpha chain | A9WR53 | 96.92% | PM0084024 |
| 135 | Tyrosine–tRNA ligase | A9WQH9 | 96.14% | PM0084062 |
| 136 | UDP-galactopyranose mutase | A9WMM1 | 95.45% | PM0084063 |
| 137 | UDP-galactose 4-epimerase | A9WN76 | 95.46% | PM0084064 |
| 138 | UDP-N-acetylglucosamine 2-epimerase | A9WMK3 | 96.44% | PM0084065 |
| 139 | Undecaprenyl-PP-MurNAc-pentapeptide-UDPGlcNAc GlcNAc transferase | A9WRD8 | 95.40% | PM0084066 |
| 140 | UDP-N-acetylmuramate–L-alanine ligase | A9WRD7 | 95.82% | PM0084067 |
| 141 | Uracil-DNA glycosylase | A9WM93 | 96.93% | PM0084068 |
| 142 | Urocanate hydratase | A9WN96 | 95.44% | PM0084069 |
Protein Ligand Docking
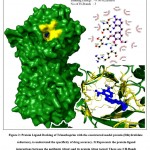
To demonstrate the application of the constructed homology models, protein-ligand study (to screen for an effective antibiotic against R. salmoninarum) was performed. Three known antibacterial drugs i.e., Isoniazid, Trimethoprim and Sulfadiazine, were subjected to protein ligand docking with their reported drug targets from R. salmoninarum (constructed in this study) as well as from other template organisms (as crystal structures from the PDB website, belonging to Mycobacterium tuberculosis, Streptococcus pneumoniae, & Burkholderia cenocepacia respectively).15,16 As shown in Table.2, the results of the docking study demonstrated that among the three antibiotics, trimethoprim exhibited the highest potential to be an effective inhibitor of the dihydrofolate reductase protein, with a binding energy of -9.06 Kcal/mol, while,Isoniazid exhibited an inconsequential binding energy of -5.36 Kcal/mol and sulfadiazine exhibited a binding energy of -6.65 Kcal/mol. The graphical representation of the protein-ligand interactions between the antibiotic (drug) and its protein (drug target) is shown in Fig.2. Among the three antibiotics, on account of it exhibiting a high binding energy of -9.06 Kcal/mol with the formation of 3 hydrogen bonds (Thr-84, Arg-85, Asn-136) and having 13 hydrophobic interactions, Trimethoprim may be proposed as a drug of choice against R. salmoninarum and the Bacterial Kidney Disease (BKD). The protein models built in this study could be applied to other docking studies as well as advanced molecular dynamic simulation studies for the purpose of recognition of effective antibiotic drugs against this specific pathogen/disease.
Table 2: Binding energies of the drug targets from R. salmoninarum and template organisms, docked against established antibiotic agents. Trimethoprim exhibited the highest potential and thus, is an effective inhibitor of the dihydrofolate reductase protein, with a binding energy of -9.06 Kcal/mol. Isoniazid exhibited an insignificant binding energy of -5.36 Kcal/mol and sulfadiazine exhibited a binding energy of -6.65 Kcal/mol.
| Antibiotic | Protein | Organism | Binding Energy (in Kcal/mol) |
| Isoniazid | Dihydrofolate reductase | Mycobacterium tuberculosis | -5.1 |
| Renibacterium salmoninarum | -5.36 | ||
| Trimethoprim | Dihydrofolate reductase | Streptococcus pneumoniae | -8.01 |
| Renibacterium salmoninarum | -9.06 | ||
| Sulfadiazine | Dihydropteroate synthetase | Burkholderia cenocepacia | -4.77 |
| Renibacterium salmoninarum | -6.65 |
Conclusion
This study purported to build 3D computational protein structures of the under-exploited R. salmoninarum which is responsible for the Bacterial Kidney Disease (BKD) of the commercially viable Salmonid fishes. The results of the study also suggests that among the test antibiotics screened, since trimethoprim demonstrated significant affinity against its drug target, it can be a potent antibiotic effective against R. salmoninarum. This proposal can be investigated further via computational approaches such as MD simulation and/or in-vitro screening of antibiotics against the said pathogen.
The structures thus developed in this study can also be helpful in understanding the pathogenesis of this disease. These can also be used to lay out a roadmap for discerning the effect of inhibition of selective proteins. Those proteins might be efficient antibiotic drug targets, and they might lead to the construction of other antibiotic chemicals that may be better than currently available drugs. This docking study offers a novel grassroots level platform for eventual in-silico as well as wet lab work on Renibacterium salmoninarum specific drug discovery, development and design.
Acknowledgement
The authors are grateful to the management of St. Joseph’s College (Autonomous), Bengaluru, Karnataka, India for supporting this piece of research work.
Conflict of Interest
The authors declare no conflict of interest.
Funding Sources
There are no funding source.
References
- Knapp G, Guettabi M, Goldsmith OS. The Economic Importance of the Bristol Bay Salmon Industry. 2013;(April):86
- Poblete EG, Drakeford BM, Ferreira FH, Barraza MG, Failler P. The impact of trade and markets on Chilean Atlantic salmon farming. Aquac Int. 2019;27(5):1465-1483. doi:10.1007/s10499-019-00400-7
CrossRef - Liu Y, Sumaila UR. An Analysis of the Management and Economics of Salmon Aquaculture. Resour Manag Environ Stud. 2008;Ph.D.(January):157.
- Sanders E, Fryer JL. Causative Agent of Bacterial Kidney Disease in Salmonid Fishes ? 1980;(5128):496-502.
CrossRef - Brynildsrud O, Feil EJ, Bohlin J, et al. Microevolution of renibacterium salmoninarum: Evidence for intercontinental dissemination associated with fish movements. ISME J. 2014;8(4):746-756. doi:10.1038/ismej.2013.186
CrossRef - David Brunoa, Bertrand Colleta, Anna Turnbulla, Rachel Kilburna, Amanda Walkera, Daniel Pendreya, Alison McIntosha, Katy Urquharta G. Evaluation and development of diagnostic methods for Renibacterium salmoninarum causing bacterial kidney disease (BKD) in the UK. Aquaculture. Volume 269(Issues 1–4):Pages 114-122. doi:https://doi.org/10.1016/j.aquaculture.2007.04.057
CrossRef - Bruno D. The relationship between auto-agglutination, cell surface hydrophobicity and virulence of the fish pathogen Renibacterium salmoninarum. FEMS Microbiol Lett. 1988;51(2-3):135-139. doi:10.1016/0378-1097(88)90387-4
CrossRef - Sanders E, Fryer JL. Renibacterium salmoninarum gen. nov., sp. nov., the Causative Agent of Bacterial Kidney Disease in Salmonid Fishes. Int J Syst Bacteriol. 1980;30(5128):496-502.
CrossRef - Kaattari SL, Piganelli JD, Physiology F. Organism , Pathogen , and Environment. In: Fish Physiology. ; 1996.
CrossRef - Kusser W, Fiedler F. Murein type and polysaccharide composition of cell walls from Renibacterium salmoninarum. FEMS Microbiol Lett. 1983;20(3):391-394. doi:10.1111/j.1574-6968.1983.tb00154.x
CrossRef - Grayson TH, Cooper LF, Wrathmell AB, Roper J, Evenden AJ, Gilpin ML. Host responses to Renibacterium salmoninarum and specific components of the pathogen reveal the mechanisms of immune suppression and activation. Immunology. 2002;106(2):273-283. doi:10.1046/j.1365-2567.2002.01420.x
CrossRef - Bruno DW, Ellis AE. Salmonid disease management. Princ Salmon Cult. Published online 1996:759-832.
CrossRef - K ASB, Imran S, Ravi L. Elucidation of Computational 3D Models of Protein Drug Targets for Colletotrichum Falcatum a Fungal Plant Pathogen Causing Red Rod of Sugarcane. Biomed Pharmacol J. 2020;13(2):1-16.
CrossRef - Padmashree AP, Imran S, Prakash T, Ravi L. Construction of 3D Model of Protein Drug Targets for Erysiphe necator a Fungal Plant Pathogen Causing Powdery Mildew. Biomed Pharmacol J. 2020;13(3):1505-1511. doi:10.13005/bpj/2024
CrossRef - Ravi L, Krishnan K. A Handbook On Protein-Ligand Docking Tool: AutoDock4. Innovare J Med Sci. 2016;4(3).
- Olson GMMDSGRSHRHWEHRKBAJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. Published online 1999.

This work is licensed under a Creative Commons Attribution 4.0 International License.