How to Cite | Publication History | PlumX Article Matrix
Identification of Pathogenic Microbes in Tools of Beauty Salon in Jeddah City
Najwa Alharbi* and Hanan Mohammed Alhashim
and Hanan Mohammed Alhashim
King Abdelaziz University, Science College, Biology Department, Jeddah, Saudi Arabia
Corresponding Author E-mail: nmaalharbi@kau.edu.sa
DOI : http://dx.doi.org/10.13005/bbra/2956
ABSTRACT:
Beauty salons may draw in customers with glamour; however, they could also be considered a major health issue. They can cause the spread of bacterial and fungal infections. The purpose of this research was to identify pathogenic microbes from beauty salon tools. Microorganisms from contaminated salon tools and cosmetic products were isolated using various selective media. Microbial isolates were identified based on their molecular and biochemical characteristics. The most common bacterial species isolated were Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus equorum, Microbacterium spp., Bacillus siamensis, Bacillus subtilis, Sphingomonas aeria, Macrococcus spp., Microbacterium oxydans, Brachybacterium spp., Micrococcus luteus, and Brachybacterium nesterenkovii. Fungal isolates included Penicillium spp., Aspergillus niger, Purpureocillium lilacium, and Aspergillus flavus. Overall, Staphylococcus spp. and A. niger were the most common organisms isolated from the samples. The presence of potential pathogens indicates that the tools used in salons have not been adequately sterilized and the high risk of diseases spread.
KEYWORDS: Beauty Salons; Pathogenic Microorganisms; Salon Product; Salons’ Tool
Download this article as:| Copy the following to cite this article: Alharbi N, Alhashim H. M. Identification of Pathogenic Microbes in Tools of Beauty Salon in Jeddah City. Biosci Biotech Res Asia 2021;18(4). |
| Copy the following to cite this URL: Alharbi N, Alhashim H. M. Identification of Pathogenic Microbes in Tools of Beauty Salon in Jeddah City. Biosci Biotech Res Asia 2021;18(4). Available from: https://bit.ly/30HGykw |
Introduction
A beauty salon’s aim is glamour—a business that makes used of a variety of tools and devices to improve the appearance of one’s hair, skin and body. Beauty products are mostly blends of chemical compounds from natural (such as coconut oil) or engineered sources 1. Within the United States, the Food and Drug Administration (FDA), which regulates the beauty care products industry, characterizes beauty care products as ” The FDA defines a cosmetic as a product (excluding pure soap) intended to be applied to the human body for cleansing, beautifying, promoting attractiveness, or altering the appearance” 2.
Cosmetic items and tools are favorable environments for the reproduction of viral, parasitic, and bacterial organisms, which contribute to and cause the spread of infections 3,4. Various components contribute to this issue. First, the components of most cosmetic products, such as organic and inorganic compounds, moisturizers, basic minerals, and growth factor such as some vitamins can provide an environment conducive to the reproduction of organisms 5,6. Second, the dates of production and expiration are, for the most part, not checked for beauty care products; thus, the decrease in effectiveness of the preservatives within the makeup over time is not noticed 7. Third, makeup is not produced under sterile conditions and is habitually shared in beauty salons 8. Fourth, the customary apparatuses used in nail salons such as clippers, scissors, and nail care instruments can inadvertently pierce the skin, which may lead to health issues ranging from inflamed skin to hepatitis 9. Service providers themselves are vulnerable to transmitting diseases among their 10,11,12. Some studies have already been undertaken to investigate whether the transmission of infections, microbes, ringworm, or parasites is related to cleaning methods in beauty salons 3.
A few bacterial species of the Streptococcus, Staphylococcus, and Pseudomonas genera are considered a major concern because they are related to numerous common infections and can cause respiratory issues and anti-microbial resistant infections owing to their pathogenic nature 7,13,14,15. A real-life case was described of a person who developed a methicillin-resistant Staphylococcus aureus (MRSA) infection after going to a beautician in London, UK 16. In 2006, in Rivers State, Nigeria, unhygienic devices led to HIV contamination and hepatitis, which are blood-borne illnesses 17. S. aureus has also caused outbreaks among salon clients in the United States18.
In general, most cosmetics brushes and other beauty tools, after being completely sanitized, still pose a risk in terms of bacterial transmission and contamination each time they come in contact with breaks in the skin 3,11,12,16. Four bacterial types are considered by pharmacopeia within the United States as indicators of contaminated tools and cosmetic products: Staphylococcus aureus, Escherichia coli, Salmonella spp., and Pseudomonas aeruginosa
The purpose of this research was to isolate and identify pathogenic microorganisms from cosmetic tools and products to enhance public awareness of the possibility of transmission of pathogenic microbes and diseases through the common products and items utilized in beauty salons.
Materials and Methods
Media Used for Microbial Isolation
A-Bacteria Isolation
Three types of media were used:
Nutrient agar
Nutrient agar was utilized for the segregation of numerous fungi and bacteria because it contains many nutrients needed for growth. One liter of NA contains yeast extract (2.0 g), NaCl (5.0 g), beef extract (1.0 g), agar (15.0 g), and peptone (5.0 g). The final pH is 7.4 ± 0.2 at 25°C.
Blood agar
Blood agar was used as a differential and selective medium for the cultivation of pathogenic organisms that cause hemolysis of the blood; one liter consisted of sheep blood (50.0 g) and blood agar base (950 g) that consists of pantone (10.0 g), NaCl (5.0 g), agar (15.0 g), cornstarch (1.0 g), and tryptic digest of beef heart (3.0 g); final pH 7.4 ± 0.2 at 25ºC.
MacConkey Agar
MacConkey Agar was utilized as a differential and specific medium for the identification of gram-negative bacteria that ferment lactose from gram-negative bacteria that do not ferment lactose. It consists of peptone (17 g), protease peptone (3 g), agar (15 g), toluylene red (0.03 g), methyl violet (0.001 g), NaCl (5 g), and 1000 ml distilled water; final pH 7.1 ± 0.2 at 25°C 19.
B-Fungi Isolation
Sabouraud dextrose agar (SDA)
Sabouraud dextrose agar (SDA) consists of peptones (10.0 g), agar (15.0 g), dextrose (40.0 g), and a final pH of 6±0.2 at 25°C 20.
Collection of Samples
Samples of cosmetic products and tools were collected with the help of the municipality of Jeddah from 16 beauty salons in different areas: south, east, and north (Table 1), especially salons where it was suspected that hygiene protocols were not closely adhered to. The collected samples were: hair dryers, eyeshadow, beauty blender, lipstick, scissors, mascara, foundation cream, eyeliner, nail care tools, combs, concealer, makeup brushes, and wax (Table 1).
Table 1: Salon regions and list of samples collected from each salon in Jeddah city.
| Salon Area | Samples | Salons# |
|
South area
|
Wax, mascara, comb, eyeliner | Salon1 |
| Lipstick, Nail Care Tools, eyeliner | Salon2
|
|
| Comb, Scissors, Dryer | Salon3 | |
| Eyeshadow, foundation Cream, eyeliner | Salon4 | |
| Comb, beauty blender | Salon5 | |
| Nail Care Tools, Makeup Brushes | Salon6 | |
| Scissors, Dryer, Makeup Brushes | Salon7 | |
| Mascara, eyeshadow, beauty blender | Salon8 | |
|
North area |
Mascara, Makeup Brushes | Salon9 |
| eyeshadow, Concealer | Salon10 | |
| Scissors, beauty blender | Salon11 | |
| Comb, Dryer | Salon12 | |
| Nail Care Tools | Salon13 | |
| East area | Lipstick, Concealer | Salon14 |
| Cream foundation, Makeup Brushes, eyeliner | Salon15 | |
| Nail Care Tools | Salon16 |
Handling of Samples
Some samples were taken whole to the laboratory because of their small size, such as beauty blenders, lipsticks, scissors, and concealers. They were placed separately in sterile plastic bags (to avoid contamination) with a code pasted on each bag to clarify the sample’s name, date of collection, and place from which it was taken.
Other samples had larger sizes, or they were the property of the salons, so a swab was taken instead, and the item left at the salon. This included wax, dryers, eyeshadow, combs, makeup brushes, mascara, foundation cream, eyeliner, and nail care tools. A swab was taken with a sterile swab stick, which was first moistened in normal saline. After each swab was taken, the swabs were kept in plastic bags and labeled appropriately. All samples were transported to the laboratory directly for microbiological analysis and kept at room temperature.
Microbial Isolation
A swab was taken with moistened sterile swab sticks from each whole item that had been taken to the laboratory like beauty blenders, lipsticks, scissors, and concealers. Then 10ml of sterile saline was added to each swab to make a dilute solution and mixed in the vortex for 10 minutes. Nine milliliters of saline solution and 1 ml of the sample solution were added into a new test tube to make 1-10 dilutions. Dilutions were made from 1/10 dilution, 1/100, 1/1000, 1/10000, and 1/100000.
Other items had been swabbed and left at the salon, such as wax, dryers, eyeshadow, combs, makeup brushes, mascara, foundation cream, eyeliner, and nail care tools. Ten milliliters of sterile saline was added to each swab to make a dilute solution and mixed in the vortex for 10 minutes. Nine milliliters of saline solution and 1 ml of the sample solution were added to a new test tube to make 1-10 dilutions. Beginning with this 1-10 dilution, the following dilutions were made: 1/100, 1/1000, 1/10000, and 1/100000.
Inoculation
Each medium previously described was inoculated with 0.1 ml of 105 dilution using a sterile pipette. The plates were incubated at 37°C for 24 h for bacteria, while the fungal plates were incubated at 25°C for 5-7 days.
After the incubation period, the total count of colonies was determined as a unit per milliliter of cosmetics (CFU ml-1). Each colony was then sub cultured on a nutrient agar dish. This step was repeated three times for each colony to confirm the purity of each isolated microbe. After that, each microbe was inoculated in a slant agar tube and preserved at 4°C, and the broth culture was mixed with 30% glycerol and stored at -80°C for long-term preservation.
Microbial Identification
Identification of Bacteria
A-Biochemical tests
Each bacterium was identified according to micromorphology characteristics such as Gram staining, colony appearance on agar plates, cell shapes under the microscope, and biochemical tests, such as catalase, oxidases, and analytical profile index (API-20E kit). The kits contain up to 20 biochemical tests, including β-galactosidase enzyme, arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, citrate test, H2S production, urea hydrolysis, tryptophan deaminase, indole test, Voges-Proskauer test, gelatinase, and fermentation of sugars (glucose, mannose, inositol, sorbitol, rhamnose, sucrose, melibiose, amygdalin, and arabinose).
B-Molecular Identification
In addition to biochemical tests, identification was carried out based on molecular aspects (16 srRNA sequences) using the QIAamp DNA mini kit. This process was accomplished in four steps: DNA isolation, PCR amplification, gel electrophoresis, and sequencing.
First, DNA was isolated from each bacterium according to the manufacturer’s instructions. Subsequently, we measured the concentration and purity of each DNA sample using Nanodrop.
Secondly, we used polymerase chain reaction (PCR). The DNA that contained the target sequence was subjected to PCR amplification using the universal primer 1100R (GGGTTGCGCTCGTTG) and 27F (AGAGTTTGATCMTGGCTCAG). This was performed using a thermal cycler with denaturation at 95°C for 5 min, annealing at 58°C for one minute, and elongation at 72°C for two minutes.
Third, the DNA fragments were separated according to their size using 1% agarose gel electrophoresis (one gram of agarose was added to 100 ml of 1X TAE buffer and microwaved for 30 s). The mixture was allowed to cool, and ethidium bromide (4 uL) was added and transferred to a tape tray. The comb was then placed for 20 minutes to set. Each sample was mixed with 5 μl of dye and placed into the wells with a 1 kb ladder and run at 100 V for 40 minutes. Finally, the gel was observed under UV light irradiation.
For sequencing analysis, the 16srRNA gene was sent to Macrogen, Korea. The BLAST database was used for the analysis of sequences in the GenBank databases [21] and confirmed by biochemical tests.
Identification of Fungi
A-Morphological identification
Fungi were identified based on their morphology using 10–40X magnification after staining with lactophenol cotton blue (LPCB).
B-Molecular Identification
Fungi were identified and classified based on molecular ITS regions. Each fungal isolate was cultured in SDAand incubated at 25°C for 5-7 days. DNA was isolated from each sample using a QIAamp DNA mini kit following the manufacturer’s instructions.
During PCR amplification, two primers were used: a reverse primer (5TCC TCC GCT TAT TGA TAT GC 3), and ITS forward primer (5GACACTCAAACAGGTGTACC3) to amplify the internal ITS regions of fungal DNA using PCR with the same setup used for bacterial amplification. After that, a 1% agarose gel was prepared and run as previously described.
Finally, for DNA sequencing, ITS regions were sent to Macrogen Company (South Korea). The DNA sequences of the fungal isolates were then used for BLAST analysis [21]. The results of the analysis were matched with the fungal isolate images under a microscope.
RESULTS
Among the 38 samples collected from different salons in Jeddah, only two samples were not contaminated: the dryer and one sample of eyeliners, while the rest of the samples were contaminated. Tables 2 and 3 indicate that some samples, such as the combs and nail care tools, showed microbial contents in all types of media used in this study. Some samples showed contamination only in one medium, such as the wax, dryer III, scissors I, scissors II, and eyeliner IV, which were grown only in the SDA medium. In addition, liquid lipstick I, foundation cream I, and foundation cream II showed contamination only in the blood agar. However, beauty blender Ⅰ showed contamination only in nutrient agar (Tables 2, 3; Figures 1,2).
Table 2: Microbial contamination in the samples from cosmetics tools.
| sample | No. of sample collected | Sample
number |
Type of media used | |||
| Nutrient agar | Blood agar | MacConkey agar | SDA | |||
| Comb | 4 | Comb Ⅰ | + | + | + | + |
| Comb Ⅱ | + | + | + | + | ||
| Comb Ⅲ | + | + | + | + | ||
| Comb Ⅳ | + | + | + | + | ||
| Makeup Brushes | 4 | Makeup B Ⅰ | + | – | + | + |
| Makeup B Ⅱ | – | + | + | + | ||
| Makeup B Ⅲ | – | + | + | + | ||
| Makeup B Ⅳ | + | – | – | + | ||
| Wax | 1 | Wax Ⅰ | – | – | – | + |
| Scissors | 3 | Sci Ⅰ | – | – | – | + |
| Sci Ⅱ | – | – | – | + | ||
| Sci Ⅲ | + | + | + | + | ||
| Dryer | 3 | D Ⅰ | – | – | – | – |
| D Ⅱ | – | – | – | – | ||
| D Ⅲ | – | – | – | + | ||
| nail care tools | 4 | FNC Ⅰ | + | + | + | + |
| FNC Ⅱ | + | + | + | + | ||
| FNC Ⅲ | + | + | + | + | ||
| FNC Ⅳ | + | + | + | + | ||
+ means there is microbial growth on the plate; – means there is no microbial growth on the plate.
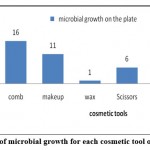 |
Figure 1: Frequency of microbial growth for each cosmetic tool obtained from salons |
Table 3: Microbial contamination in the samples from cosmetic products.
| sample | No. of sample collected | Sample number | Type of media used | |||
| Nutrient agar | Blood agar | MacConkey agar | SDA | |||
| mascara | 3 | mascara Ⅰ | + | + | – | – |
| mascara Ⅱ | + | + | + | – | ||
| mascara Ⅲ | – | + | + | – | ||
| eye shadow | 3 | E shadow Ⅰ | + | – | + | – |
| E shadow Ⅱ | + | + | + | – | ||
| E shadow Ⅲ | – | + | – | – | ||
| Liquid lipstick | 2 | Liquid lipstick Ⅰ | – | + | – | + |
| Liquid lipstick Ⅱ | + | – | + | + | ||
| Concealer | 2 | Concealer Ⅰ | + | + | + | + |
| Concealer Ⅱ | + | + | + | + | ||
| Eyeliner | 4 | Eyeliner Ⅰ | – | + | + | + |
| Eyeliner Ⅱ | + | + | + | _ | ||
| Eyeliner Ⅲ | – | – | – | _ | ||
| Eyeliner Ⅳ | – | – | – | + | ||
| Foundation
Cream |
2 | Foundation Cream Ⅰ | – | + | – | – |
| Foundation Cream Ⅱ | – | + | – | – | ||
| Beauty Blender | 3 | Beauty Blender Ⅰ | + | – | – | – |
| Beauty Blender Ⅱ | + | + | + | – | ||
| Beauty Blender Ⅲ | + | – | + | – | ||
+ means there is microbial growth on the plate; – means there is no microbial growth on the plate.
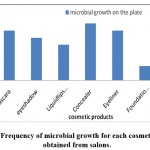 |
Figure 2: Frequency of microbial growth for each cosmetic product obtained from salons. |
Total Count of Colonies
Tables 4 and 5 show the total count of colonies isolated from each of the salon samples as a unit per milliliter of cosmetics (CFU/ ml). The tables indicated that nail tools, a comb, and scissors were contained the most microbes; while the least samples that contained microbes were Liquid lipstick.
Table 4: Bacterial counts (10⁵ CFU/ ml) in samples collected from all salons.
| sample | number of the plates | total | average | 10⁵ CFU/ ml |
| comb | 12 | 257 | 21.4 | 21.4 X10⁵ |
| Makeup brushes | 7 | 117 | 16.7 | 16.7 X10⁵ |
| Scissors | 3 | 280 | 93.3 | 93.3 X10⁵ |
| nail care tools | 12 | 377 | 31.4 | 31.4 X10⁵ |
| mascara | 7 | 144 | 20.5 | 20.5 X10⁵ |
| eye shadow | 6 | 47 | 7 | 7 X10⁵ |
| Liquid lipstick | 3 | 48 | 16 | 16 X10⁵ |
| Concealer | 6 | 215 | 35.8 | 35.8 X10⁵ |
| Eyeliner | 5 | 63 | 12.6 | 12.6 X10⁵ |
| Foundation Cream | 2 | 86 | 43 | 43 X10⁵ |
| Beauty Blender | 6 | 107 | 17.8 | 17.8 X10⁵ |
Table 5: Fungal counts ( 10⁵ CFU/ ml) in samples collected from a salon.
| sample | number of the plates | total | average | 10⁵ CFU/ ml |
| comb | 4 | 78 | 19.5 | 19.5 X10⁵ |
| Makeup brushes | 4 | 65 | 16.3 | 16.3 X10⁵ |
| Scissors | 3 | 12 | 4 | 4 X10⁵ |
| nail care tools | 4 | 55 | 13.7 | 13.7 X10⁵ |
| wax | 1 | 7 | 7 | 7 X10⁵ |
| Liquid lipstick | 2 | 5 | 2.5 | 2.5 X10⁵ |
| Concealer | 2 | 17 | 8.5 | 8.5 X10⁵ |
| Eyeliner | 2 | 25 | 12.5 | 12.5 X10⁵ |
| Dryer | 1 | 8 | 8 | 8 X10⁵ |
Microbial identification
Bacteria
Bacterial segregates were identified according to the biochemical tests shown in Table 6. The test results were compared to Bergey’s manual, which revealed the following microbes: Staphylococcus aureus, Microbacterium spp., Staphylococcus epidermidis, Bacillus siamensis, Sphingomonas aeria, Staphylococcus equorum, Macrococcus spp., Microbacterium oxydans, Brachybacterium spp., Micrococcus luteus Brachybacterium nesterenkovii, and Bacillus subtilis.
Table 6: Biochemical tests performed using API-20E kit, gram staining, catalase test, and oxidase test of each bacterium obtained from beauty salons.
| gram staining | catalase test, | oxidase test | citrate test | indole test | H2S production | urea test | sugar fermentation test | Gelatin hydrolysis test | organism |
| +ve | +ve | – ve | +ve | -ve | -ve | +ve | +ve | +ve | Staphylococcus aureus |
| +ve | +ve | -ve | +ve | +ve | +ve | -ve | -ve | +ve | Microbacterium spp |
| +ve | +ve | -ve | +ve | -ve | +ve | +ve | +ve | -ve | Staphylococcus epidermidis |
| +ve | -ve | +ve | +ve | +ve | -ve | +ve | +ve | -ve | Bacillus siamensis |
| +ve | -ve | +ve | +ve | +ve | +ve | -ve | +ve | -ve | Sphingomonas aeria |
| +ve | +ve | +ve | +ve | +ve | -ve | +ve | – ve | +ve | Staphylococcus equorum |
| +ve | +ve | +ve | -ve | +ve | -ve | -ve | -ve | +ve | Macrococcus spp |
| +ve | +ve | -ve | -ve | +ve | -ve | -ve | -ve | -ve | microbacterium oxydans |
| +ve | +ve | +ve | +ve | +ve | +ve | +ve | +ve | +ve | Brachybacterium spp |
| Variable | +ve | -ve | +ve | -ve | +ve | +ve | -ve | +ve | Micrococcus luteus |
| +ve | +ve | -ve | +ve | +ve | +ve | -ve | -ve | -ve | Brachybacterium nesterenkovii |
| +ve | +ve | Variable | -ve | -ve | -ve | +ve | -ve | +ve | Bacillus subtilis |
+ve means positive test; -ve means negative test.
Morphological Identification of Fungi
Four types of fungi were morphologically identified after staining with lactophenol cotton blue (LPCB) and examined under a light microscope (Figure 3). In addition, fungal colonies were shaped on the SDA culture media (Figure 4). The fungal isolates were Purpureocillium lilacinum, Aspergillus flavus, Penicillium spp., and Aspergillus niger.
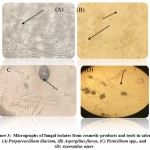 |
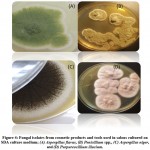
Figure 3: Micrographs of fungal isolates from cosmetic products and tools in salons; (A) Purpureocillium lilacium, (B) Aspergillus flavus, (C) Penicillium spp., and (D) Aspergillus niger |
Molecular Identification of Bacteria and Fungi
The resulting sequences of 16 srRNA of bacteria and the ITS4 of fungi were aligned in the NCBI database, and the matching sequences were identified, supported by the biochemical tests that had been done previously. Based on the above, the bacterial isolates from cosmetic products and tools were Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus equorum, Microbacterium spp., Bacillus siamensis, Bacillus subtilis, Sphingomonas aeria, Macrococcus spp., Microbacterium oxydans, Brachybacterium spp., Micrococcus luteus, and Brachybacterium nesterenkovii. The fungal isolates were Aspergillus niger, Aspergillus flavus, Penicillium spp., and Purpureocillium lilacium. Table 8 indicates that bacterial isolates were more commonly found than fungal isolates. In addition, Staphylococcus aureus, Staphylococcus equorum, Sphingomonas aeria, and Aspergillus niger were the most common organisms isolated from samples, as shown in Table 8 and Figures 6 and 7.
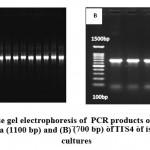 |
Figure 5: Agarose gel electrophoresis of PCR products of (A) 16S rRNA gene of bacteria (1100 bp) and (B) (700 bp) of ITS4 of isolated fungal cultures |
Table 7: The matching ratio of the alignment of bacteria and fungi isolates in the NCBI database of each microbe obtained from beauty salons.
|
Match ratio |
Organism |
| 99% | Staphylococcus aureus |
| 96% | Microbacterium spp |
| 99% | Staphylococcus epidermidis |
| 99% | Bacillus siamensis |
| 97% | Sphingomonas aeria |
| 98% | Staphylococcus equorum |
| 99% | Macrococcus spp |
| 95% | microbacterium oxydans |
| 99% | Brachybacterium spp |
| 99% | Micrococcus luteus |
| 99% | Brachybacterium nesterenkovii |
| 99% | Bacillus subtilis |
| 95% | Aspergillus niger |
| 98% | Aspergillus flavus |
| 99% | Penicillium spp. |
| 99% | Purpureocillium lilacium |
Table 8: Bacterial and fungal isolates obtained among cosmetic products and tools.
| Samples | Bacterial isolates | Fungal isolates |
| comb | Staphylococcus aureus, Microbacterium spp., Staphylococcus epidermidis, Bacillus siamensis | Aspergillus niger, Aspergillus flavus |
| Makeup brushes | Staphylococcus aureus, Sphingomonas aeria | Aspergillus niger, Aspergillus flavus |
| Scissors | Staphylococcus aureus, Staphylococcus equorum, Macrococcus spp.,
|
Penicillium spp. |
| nail care tools | Staphylococcus equorum, micro bacterium oxydans, Brachybacterium spp. Macrococcus spp, Bacillus siamensis, Sphingomonas aeria | Aspergillus niger, Aspergillus flavus |
| wax | – | Penicillium spp. |
| Dryer | – | Penicillium spp. |
| mascara | Staphylococcus aureus, Micrococcus luteus, Staphylococcus equorum, Brachybacterium nesterenkovii, Sphingomonas aeria | – |
| eye shadow | Staphylococcus aureus, Bacillus subtilis, Staphylococcus equorum, | – |
| Liquid lipstick | Staphylococcus equorum., Staphylococcus epidermidis | Aspergillus niger |
| Concealer | Staphylococcus aureus., Brachybacterium nesterenkovii, Bacillus siamensis, Sphingomonas aeria | Aspergillus niger |
| Eyeliner | Staphylococcus equorum, Macrococcus spp, Sphingomonas aeria | Purpureocillium lilacium |
| Beauty Blender | Staphylococcus epidermidis, Brachybacterium nesterenkovii, Macrococcus spp, Sphingomonas aeria | – |
| Foundation
Cream |
Micrococcus spp., | – |
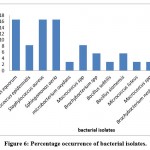 |
Figure 6: Percentage occurrence of bacterial isolates. |
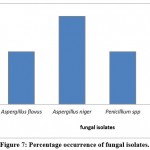 |
Figure 7: Percentage occurrence of fungal isolates. |
Discussion
This study aimed to increase awareness of diseases that may be transmitted through popular items or products utilized in beauty salons. Considering the results of this investigation, we hope that the standards of care applied in salons concerning the sterilization of beauty tools and products, and adequate storage, will be improved. These recommendations will contribute significantly to preventing the spread of infections via beauty salons.
Cosmetic tools and items are ideal environments for the proliferation of microbes; thus, they may contribute to the spread of various diseases 3,22. All but two of the samples in this study, which were obtained from multiple salons in Jeddah, Saudi Arabia, were found to be contaminated with bacteria and/or fungi. Specifically, more bacterial than fungal species were isolated from cosmetic products, which was most likely due to the more effective anti-fungal activity of preservative compounds used in cosmetic products 23. Several studies have examined the presence of an assortment of microorganisms in beauty salon products and items to highlight diseases that can be transmitted through them. For example, Ebuara et al. (2020) 24 collected and analyzed samples from clippers, clipper steps, combs, and brushes from 40 different beauty salons in Taraba State, Nigeria, which were all found to contain pathogenic bacteria, including Staphylococcus, Bacillus, and Streptococcus, as well as pathogenic fungi of the Aspergillus, Trichophyton, Malasseza, Mucor, and Microsporum genera.
In this study, the bacterial isolates from cosmetic products and tools were Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus equorum, Microbacterium spp., Bacillus siamensis, Bacillus subtilis, Sphingomonas aeria, Macrococcus spp., Microbacterium oxydans, Brachybacterium spp., Micrococcus luteus, and Brachybacterium nesterenkovii. The fungal isolates were Aspergillus niger, Aspergillus flavus, Penicillium spp., and Purpureocillium lilacium. Our results agree with those of several authors who isolated different species of bacteria and fungi from the tools and products used in a salon 4,7,9,22,24,25,26,27,28,29,30,31.
In this study, the highest microbial contents were observed in the comb and nail care kits, among other tools used in cosmetics. These tools were found to be contaminated with S. aureus, S. epidermidis, S. equorum, Microbacterium oxydans, Bacillus siamensis, Brachybacterium spp., Macrococcus spp., S. aeria, Microbacterium spp., Aspergillus niger, and Aspergillus flavus. Therefore, it can be inferred that these apparatuses received the lowest level of sterilization of all the tested devices. Additionally, nail care devices can inadvertently pierce the skin, which may lead to health issues ranging from inflamed skin to hepatitis 9. Our results showed that, among all the tested cosmetic products, mascaras had increased bacterial variety, including S. aureus, M. luteus, S. equorum, B. nesterenkovii, and S. aeria. This is most likely due to its hydrous structure, which makes it a more favorable environment for the proliferation of eye-infection causing pathogens. In a similar study, Dadashi and Dehghanzadeh (2016) 4 reported that mascaras had increased bacterial diversity compared to other examined cosmetic products. In the present study, S. aureus, S. equorum, S. aeria, and A. niger were the most commonly isolated organisms in the study samples, which agrees with a study by Stanley et al.(2019) 9, in which S. aureus and A. flavus were the predominant isolates. Similarly, other studies also reported that Staphylococcus spp. was the predominant isolate among all samples obtained from salons 3.4. Both S. aureus and S. epidermidis were isolated from almost all the samples. These microbes are related to nosocomial contamination and are not effectively controlled by antibiotics 32. Some of the most frequent bacterial species identified in salon instruments and products in the present study, such as Bacillus spp., can cause food poisoning, as they can secrete toxins into food that will in turn trigger gastrointestinal illnesses 33. Additionally, the bacterial genus Brachybacterium spp. was isolated from the nail care tools; although this microbe is rarely considered a human pathogen, a recent clinical case report described an individual who had an eye infection caused by B. paraconglomeratum 34. In our study, M. luteus, a bacterium that is harmful to human health, causing skin, blood, and mucous membrane contamination, was isolated from mascara. In 2019, a similar study by Alswedi and Jaber (2019) 25 also isolated this pathogen from cosmetic tools. In contrast, some bacterial isolates from our study, such as Macrococcus spp., Microbacterium spp., and S. aeria, are not known pathogens. Nonetheless, some Microbacterium and Sphingomonas genera can cause bacteremia and several infections, including S. paucimobilis 35,36.
In addition to bacterial pathogenic isolates, fungal pathogens have been isolated from tools and products used in salons, such as Aspergillus spp., which causes aspergillosis in humans, and can secrete aflatoxin, which is a carcinogenic and toxic metabolite 37. Moreover, Penicillium spp., which was obtained from salon samples, such as wax, dryer, and scissors, can cause spoilage of some foods 38. Accordingly, several studies have also isolated Aspergillus spp. and Penicillium spp. from cosmetic tools and products 3,4,9,13,22,24,27,29. P. lilacinum was also detected in the eyeliner samples. Although this fungus rarely causes diseases in humans, it may pose a health risk to immunocompromised individuals 39.
Conclusion
The presence of potential pathogens indicates that the tools used in salons have not been adequately sterilized. It has been observed that each salon uses different sterilization techniques, with approximately 35% of service providers using ultraviolet sterilization, 20% using quartz beads, and only 1% of providers reporting the use of an ultrasonic cleaner to sanitize their tools 40. However, none of these methods has been proven adequate for achieving a satisfactory level of sterilization. Therefore, various sterilization approaches should be used.
The outcomes of this study might have been limited by the variable types of microorganisms present in the collected samples. In addition, the media and methods used may not be sufficient to isolate all contaminating microorganisms, as some microorganisms require highly specific conditions to grow, as in the case of parasites and viruses.
We recommend that salons are required to take care over the storage and sterilization methods used for beauty equipment and products. In addition, we suggest the use of personal instead of public cosmetic kits. All these suggestions may help to avoid the spread of diseases and infections through salons.
Acknowledgement
The authors would like to thank the staff of King Fahd research center for their support and cooperation during this study.
Conflict of Interest
The authors declare no conflict of interest.
Funding Sources
This work was not supported by any funding agencies.
References
- Posted by Dr Oliver Jones, Professor Ben Selinger, AM.( September 19,2019 ); The chemistry of cosmetics from Australian Academy of Science Website: https://www.science.org.au/curious/peoplemedicine/chemistry-cosmetics
- posted by U.S. Food and Drug Administration.(March1,2021) Cosmetic Products & Ingredients. Website: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm2006949.htm.)https://catalog.hardydiagnostics.com/cp_prod/Content/hugo/PotatoDextroseAgar.htm
- Enemuor, S., Ojih, M., Isah, S., & Oguntibeju, O. (2013). Evaluation of bacterial and fungal contamination in hairdressing and beauty salons.
CrossRef - Dadashi, L., & Dehghanzadeh, R. (2016). Investigating incidence of bacterial and fungal contamination in shared cosmetic kits available in the women beauty salons. Health promotion perspectives, 6(3), 159.
CrossRef - Rope, B. L. (2002). Conquering contamination. Global cosmetic industry, 170(1), 40-43.
CrossRef - Pinon, A., Alexandre, V., Cupferman, S., Crozier, A., & Vialette, M. (2007). Growth, survival, and inactivation of Pseudomonas aeruginosa and Staphylococcus aureus strains of various origin in the presence of ethanol. International journal of cosmetic science, 29(2), 111-119.
CrossRef - Behravan, J., Bazzaz, F., & Malaekeh, P. (2005). Survey of bacteriological contamination of cosmetic creams in Iran (2000). International journal of dermatology, 44(6), 482-485.
CrossRef - Giacomel, C., Dartora, G., Dienfethaeler, H., & Haas, S. (2013). Investigation on the use of expired make‐up and microbiological contamination of mascaras. International journal of cosmetic science, 35(4), 375-380.
CrossRef - Stanley, H., Oba, T., & Ugboma, C. (2019). Evaluation of Microbial Contamination of Combs and Brushes in Beauty Salons within the University of Port Harcourt, Rivers State, Nigeria. Archives of Current Research International, 1-7.
CrossRef - De Souza, B. A., & Shibu, M. M. (2004). Infectious and respiratory hazards of nail sculpture. Plastic and reconstructive surgery, 114(4), 1004.
CrossRef - Stout, J. E., Gadkowski, L. B., Rath, S., Alspaugh, J. A., Miller, M. B., & Cox, G. M. (2011). Pedicure-associated rapidly growing mycobacterial infection: an endemic disease. Clinical infectious diseases, 53(8), 787-792.
CrossRef - Edward, S. M., Megantara, I., & Dwiyana, R. F. (2015). Detection of fungi in hair-brushes in beauty salons at Jatinangor. Althea Medical Journal, 2(4), 516-520.
CrossRef - Anelich, L., & Korsten, L. (1996). Survey of micro‐organisms associated with spoilage of cosmetic creams manufactured in South Africa. International journal of cosmetic science, 18(1), 25-40
CrossRef - Draelos, Z. D. (2001). Special considerations in eye cosmetics. Clinics in dermatology, 19(4), 424-430
CrossRef - Tharmila, S., Jeyaseelan, E. C., & Thavaranjit, A. (2012). Inhibitory effect of some traditional hair washing substances on hair borne bacteria. Der Pharmacia Lettre, 4(1), 199-204.
- Ruddy, M., Cummins, M., & Drabu, Y. (2001). Hospital hairdresser as a potential source of cross-infection with MRSA. Journal of Hospital Infection, 49(3), 225-227.
CrossRef - Pelenita TM. Acrylic Nail and Native Bacteria. Saint Martin’s University Biolog Journal. 2006;1:67-92
CrossRef - Huijsdens, X. W., Janssen, M., Renders, N. H., Leenders, A., Van Wijk, P., van Santen-Verheuvel, M. G., . . . Morroy, G. (2008). Methicillin-resistant Staphylococcus aureus in a beauty salon, the Netherlands. Emerging infectious diseases, 14(11), 1797.
CrossRef - Atlas, R. M. (2006). The handbook of microbiological media for the examination of food: CRC press.
CrossRef - Posted by Sagar Aryal.( September 13, 2018); Culture Media » Sabouraud Dextrose Agar (SDA) from Microbe Notes Website: https://microbenotes.com/sabouraud-dextrose-agar-sda/ .
- Benson, D. A., Boguski, M. S., Lipman, D. J., Ostell, J., & Ouellette, B. (1998). GenBank. Nucleic acids research, 26(1), 1-7.
CrossRef - Stanley, M. C., Ifeanyi, O. E., & Kingsley, O. (2015). Evaluation of Microbial Contamination of Tools Used in Hair Dressing Salons in Michael Okpara University of Agriculture, Umudike, Abia State, Wiszniewska, M. & Walusiak-Skorupa, J. Recent trends in occupational contact dermatitis. Current allergy and asthma reports, 15, 1-11.
- Mislivec, P., Beuchat, L., & Cousin, M. (1992). Yeasts and molds. Compendium of methods for the microbiological examination of foods, 3, 239-243.
- Ebuara, F., Imarenezor, E., Brown, S., Aso, R., Obasi, B., & Tyovenda, E. ISOLATION AND IDENTIFICATION OF PATHOGENIC MICROORGANISMS ASSOCIATED WITH BARBERS’EQUIPMENT IN WUKARI, TARABA STATE, NIGERIA
- Alswedi, F. G., & Jaber, A. S. (2019). Isolation of Pathogenic Bacteria from some Male Barbershops in the City of Nasiriyah. INTERNATIONAL JOURNAL OF PHARMACEUTICAL AND CLINICAL RESEARCH, 10(02), 233-241
CrossRef - Hassan, S. M., Hamad, A. K., Shallal, A. F., & Abdullah, S. M. (2018). Isolation of pathogenic microbes from beauty salons in Ranya, Iraq. Galen Medical Journal, 29, 104-106.
CrossRef - Janmohammadi, F., Fathi, G., Roshani, D., & Farahmandi, K. (2016). Evaluation of bacterial and fungal contaminations in barbershops in Kamyaran city, Iran-Summer 2015. INTERNATIONAL JOURNAL OF MEDICAL RESEARCH & HEALTH SCIENCES, 5(9), 368-371.
- Naz, S., Iqtedar, M., Ul Ain, Q., & Aftab, K. (2012). Incidence of Human Skin Pathogens from Cosmetic Tools used in Beauty Salons in Different Areas of Lahore, Pakistan. Journal of Scientific Research, 4(2), 523-523.
CrossRef - Onajobi IB, Okerentugba PO, Adeyemi SA Laba SA. Microbial evaluation of manicure and pedicure shops along Adewole Estate Ilorin Kwara, Nigeria. Nigerian Journal of Microbiology. 2015;28:2939-2945
- Salem, A. H., Mietzel, T., Brunstermann, R., & Widmann, R. (2018). Two-stage anaerobic fermentation process for bio-hydrogen and bio-methane production from pre-treated organic wastes. Bioresource technology, 265, 399-406
CrossRef - Sekula, S. A., Havel, J., & Otillar, L. J. (2002). Nail salons can be risky business. Archives of dermatology, 138(3), 414-415.
CrossRef - Horan, T. C., Andrus, M., & Dudeck, M. A. (2008). CDC/NHSN surveillance definition of health care–associated infection and criteria for specific types of infections in the acute care setting. American journal of infection control, 36(5), 309-332.
CrossRef - Tuazon, C. U. (2016). Bacillus species. Last accessed on.
- Murata, K., Ozawa, K., Kawakami, H., Mochizuki, K., & Ohkusu, K. (2020). Brachybacterium paraconglomeratum Endophthalmitis Postcataract Operation. Case reports in ophthalmological medicine, 2020.
CrossRef - Ryan, M., & Adley, C. (2010). Sphingomonas paucimobilis: a persistent Gram-negative nosocomial infectious organism. Journal of Hospital Infection, 75(3), 153-157
CrossRef - Toh, H.-S., Tay, H.-T., Kuar, W.-K., Weng, T.-C., Tang, H.-J., & Tan, C.-K. (2011). Risk factors associated with Sphingomonas paucimobilis infection. Journal of Microbiology, Immunology, and Infection, 44(4), 289-295.
CrossRef - Awuchi, C. G., Amagwula, I. O., Priya, P., Kumar, R., Yezdani, U., & Khan, M. G. (2020). Aflatoxins In Foods and Feeds: A Review on Health Implications, Detection, And Control. Bull. Env. Pharmacal. Life Sci, 9, 149-155.
- Kirk, P., Cannon, P., Minter, D., & Staplers, J. (2008). Dictionary of the Fungi Wallingford. UK: CABI, 335.
- Saberhagen, C., Klotz, S. A., Bartholomew, W., Drews, D., & Dixon, A. (1997). Infection due to Actinomyces lilacinus: a challenging clinical identification. Clinical Infectious Diseases, 25(6), 1411-1413.
CrossRef - Rideout, K. (2010) “Comparison of Guidelines and Regulatory Frameworks for Personal Services Establishments.” Vancouver , BC: National Collaborating Centre for Environment Health . www.ncceh.c

This work is licensed under a Creative Commons Attribution 4.0 International License.