How to Cite | Publication History | PlumX Article Matrix
Kohinoor Kaur , Samiksha Sharma
, Samiksha Sharma , Nidhi Shree
, Nidhi Shree and Rekha Mehrotra*
and Rekha Mehrotra*
Department of Microbiology, Shaheed Rajguru College of Applied Sciences for Women, Delhi University, Vasundhara Enclave, Delhi – 110096, India.
Corresponding Author E-mail:rekha.mehrotra@rajguru.du.ac.in
DOI : http://dx.doi.org/10.13005/bbra/3063
ABSTRACT: Plastic has become an indispensable part of our lives and cutting down plastic consumption entirely is difficult to achieve. The recalcitrant and non-biodegradable nature of plastic leads to accumulation of tons of plastic in landfills and water bodies which further risks marine life and human life too causing serious health issues. In recent years, several microbial enzymes have been discovered that have the ability to degrade plastic. The present review highlights the recent discovery and properties of the plastic-eating bacteria, Ideonella sakaiensis, that has potential to be used for plastic degradation and recycling. The bacteria possess unique enzymes that allow it to utilise Polyethylene terephthalate (PET) plastic, thereby degrading it to relatively safer monomeric forms that can be further degraded and purified to manufacture recycled plastics. The review focuses on the mechanism of PET hydrolysis, recent advances in the field to escalate enzymatic efficiency and development of new bacterial and enzymatic strains through genetic engineering which can enhance its catalytic competence and make the process time and cost-effective. The plastic metabolising bacteria can thus be a potential and efficient bio-alternative to degrade plastic in a biological and sustainable manner thereby helping scale the otherwise insurmountable plastic pollution crisis.
KEYWORDS: Biodegradable; Bioremediation; Ideonella; MHET; PET; Plastic
Download this article as:| Copy the following to cite this article: Kaur K, Sharma S, Shree N, Mehrotra R. Recent Advancements and Mechanism of Plastics Biodegradation Promoted by Bacteria: A Key for Sustainable Remediation for Plastic Wastes. Biosci Biotech Res Asia 2023;20(1). |
| Copy the following to cite this URL: Kaur K, Sharma S, Shree N, Mehrotra R. Recent Advancements and Mechanism of Plastics Biodegradation Promoted by Bacteria: A Key for Sustainable Remediation for Plastic Wastes. Biosci Biotech Res Asia 2023;20(1). Available from: https://bit.ly/3QvbKcR |
Introduction
Plastic pollution is the current global threat which is accelerating at a speed which if not controlled, can lead to disastrous impacts in the near future. Plastics are organic polymers, having high molecular weight and are synthesized via polymerization/polycondensation of natural material like cellulose, coal and crude oil; with carbon, hydrogen, nitrogen, oxygen, chlorine, sulphur and silicon atoms serving as its major key components. Polyethylene terephthalate (PET) is a petroleum-based plastic polymer composed of monomeric residues of terephthalic acid (TPA) and ethylene glycol (EG) bonded via ester linkages which contributes significantly to the molecule’s stability. Most percentages of daily-use disposable plastic items like drinking bottles and packets consist of PET, which due to its low cost and easy production has become a popular option amongst manufacturers[1]. However, statistics reveal that out of the hundred million tons of plastic waste generated; only a tiny fraction of it is recycled due to the inert and non-biodegradable nature of the plastic polymer. Plastic pollutants along with disrupting the environmental balance can also cause serious harm to the flora and the fauna. Microplastics are an alarming concern, which can prove to be fatal if consumed accidentally and have even invaded a pregnant woman’s womb, owing to their small size and toxic nature[2].
In order to reduce the plastic burden, various methods are being, namely, physical recycling, chemical recycling and biodegradation being the most recent one. Physical recycling involves melting of washed granules or pellets of the plastic so as to reduce the contamination, this is also known as ‘material recycling’. The basic polymer remains same throughout the process. Once melted, they can be moulded into various shapes. Since the process involves heating of the plastic, it can lead to thermal degradation of the polymer chains, and the loss of their mechanical properties. As a result, beverage bottles no longer qualify for reprocessing as beverage bottles [3,4]. Incineration of PET has been carried out in various countries to generate heat and electricity since polymers are high-yielding energy sources. However, this method has been widely accused as ecologically unacceptable owing to the health risk from air borne toxic substances, like dioxins (in the case of chlorine containing polymers)[3,5].
The chemical methods involve the synthesis of monomer unit products from the polymer backbone which is accomplished using chemical techniques[6]. Chemical recycling of PET involves hydrolysis, methanolysis, aminolysis, ammonolysis and glycolysis. Hydrolysis can be acidic, alkaline or neutral depending on the reaction conditions[7,5]. The disadvantage lies in the fact that it requires extreme temperature and pressure conditions for the reaction to be carried out. This necessitates the development of reactors capable of withstanding such extreme conditions. Further, acidic hydrolysis requires the use of harsh chemicals, such as, HNO₃ or concentrated H₂SO₄, that pose a high class of industrial hazard and risk associated with tightening environmental legislation[8]. Similar to hydrolysis, methanolysis is carried out at 180-280°C at 2-4MPa; maintaining such extreme conditions becomes less cost effective. During the process of glycolysis, EG is introduced into the PET chain to produce BHET and higher oligomers. The resulting BHET can then be employed as a substrate for the production of new PET and other oligomers. Because the products created in this process are not distinct, it is difficult to obtain a product with a higher degree of purity[5]. Furthermore, the cost of separation and refining for the products is extremely high for these methods, making it an economically undesirable option[9].
On the other hand, biodegradation can help with cleaner and gentler plastic disintegration by utilising bacteria and their enzymes. With the discovery of a novel species of the genus Ideonella, called Ideonella sakaiensis as reported by Yoshida et al., in 2016, enzymatic degradation by the bacteria can be exploited to recycle plastic. This bacterium overcomes the major drawback faced by the chemical recycling methods, i.e., it does not require extreme conditions and can function at an optimum temperature of 30-37°C and an optimum pH range of 7-7.5 at a normal atmospheric pressure[11]. These enzymes work on ester bonds to hydrolyse them and release the constituent monomers, hence, giving a higher level of purity and easier isolation of the constituent products. Since, it is carried out by microbial activity, making it a more natural form of recycling than the chemical ones[3]. Further, no harmful by-products are known to be produced during the biodegradation process. Nonetheless, more research and engineering are required to create new and improved enzymes with higher binding efficiency to PET, greater thermal stability, and hydrolytic activity[12].
When comparing biodegradation with other recycling procedures, it is clear that biodegradation is more environmentally friendly with reduced formation of toxins and aerosols. Furthermore, in comparison to biodegradation, these chemical methods necessitate more extreme conditions and lower purity of product being isolated. Since it is carried out by microbes, biodegradation is much less expensive than other prevailing methods.
Discovery and Properties of Ideonella sakaiensis
Kyoto Institute of Technology’s Kohei Oda, Keio University’s Kenji Miyamoto, and co-workers in March 2016 discovered the first known plastic-eating bacteria, ‘Ideonella sakaiensis’. The bacterium was isolated from sludge outside a bottle recycling facility in Osaka. This bacterium has gained the ability to break down PET plastic in an organic way[10]. The bacterium belongs to the class Betaproteobacteria, family Comamonadaceae, and genus Ideonella[13].
Ideonella sakaiensis, the plastic-eating bacteria has the ability to degrade PET plastics organically (as a carbon and energy source). When viewed under the microscope, the bacterial cells appear rod-shaped with dimensions of 0.6–0.8µm wide and 1.2–1.5µm long. Other tests reveal that these bacteria are non-spore formers having a gram-negative cell wall. Additionally, they are aerobic bacteria with a polar flagellum which aids in their motility. Though the bacteria grow the best at an optimum temperature range of 30-37°C and an optimum pH range of 7-7.5, they can even thrive in a temperature in the span of 15°C – 42°C and a pH of 5.5-9. The bacteria are positive for catalase and cytochrome oxidation tests. It tests positive for uptake of NAG, maltose, adipic acid and potassium gluconate and negative for denitrification, NO3- reduction, indole formation and sugar (glucose) fermentation in the API 20 NE test, β-galactosidase and D-glucose assimilation (Table 1) [14,15,11].
 |
Table 1: Depicts various characteristics of Ideonella sakaiensis along with its electron Micrographs. |
The bacterium grows well on PET agar (selective media), forming circular, colourless, smooth colonies with about 0.5-1mm in length. The bacterium has a DNA G+C content of about 70.4mol%. Phylogenetic, biological, biochemical, and physiological studies reveal the bacteria as a new representative of the genus Ideonella, named I. sakaiensis[14,15,11]. Fig. 1 depicts the phylogenetic association of the bacterium with other plastic degrading bacteria.
 |
Figure 1: Neighbourhood-joining phylogenetic tree using 16S rRNA sequences showing the relation between different plastic degrading bacterial strains. |
The bacterium can be cultured under laboratory conditions using the NRBC no. 802 agar medium. The medium consists of polypepton, yeast extract, magnesium sulphate, dipotassium hydrogen phosphate, distilled water, and agar. It has been observed that the growth of the organism is inhibited if the temperature is increased above 45°C. A concentration of 3% (w/v) NaCl salt also prevents the growth of the bacteria[2]. When the bacteria I. sakaiensis was cultured in a broth medium, almost no PET hydrolysis was detected indicating that I. sakaiensis degrades and assimilates PET to produce carbon dioxide under aerobic conditions[16]. The aerobic bacterium neither responds positively nor negatively to light. It does not possess any visible pigments either. The cells of I. sakaiensis 201-F6 adhere to the PET film during the growth and are connected to each other via appendages. It is believed that the appendages may be involved in the delivery of PET-degrading enzymes, IsPETase onto the film[16].
The isolation of I. Sakaiensis involves three steps. The first step requires the activation of the broth culture or the solid NBRC media directly from cryo culture and multiple liquid cultures. Using modified MacConkey agar, colonies are isolated, as MacConkey prevents the growth of gram-positive and I. sakaiensis is a gram-negative bacterium, the colonies obtained are streaked and re-streaked every 24 hours for obtaining an isolated colony. Samples of the bacteria obtained are tested via Polymerase Chain Reaction (PCR) to identify the species growing on the plate[11].
PET degrading ability of Ideonella sakaiensis
PET is a petroleum-based plastic which can be synthesized via polycondensation of its monomeric residues viz. TPA and EG or by transesterification of dimethyl terephthalate (DMT) and EG. PET polymers are highly inert, thermally stable, transparent, impermeable to gases and liquids and have good mechanical strength which makes them a popular choice for the production of daily-use plastic items like bottles, containers, food packets, textiles and clothing[3]. With such a great influence on our daily lives, PET accounts for more than 70% of the plastic consumption along with other plastic polymers like polyesters, polysulfates and polyamides; and about 12% of the global solid waste, posing a serious threat to the ecosystems[17].
PET plastics are very stable, chemically inactive and highly resistant to hydrolytic or microbial degradation, the key factors to which are their low chain mobility, high surface hydrophobicity and crystalline nature[8]. This crystallinity makes the degradation process more difficult for the microbes which degrade flexible amorphous solids with better ease. The highly stable nature of the C=C bonds and ester bonds makes the situation all the more critical. In spite of all these inert features, scientists have discovered various PET hydrolytic enzymes with different degradation speeds and mechanisms[11].
I. sakaiensis feeds on PET plastic as a carbon and energy source for their metabolic activities. Amongst the different types of plastics available, the bacterium has a key potential to degrade PET by hydrolyzing it into its monomeric forms which can then further be processed and recycled into new plastic. The process starts with the binding of the extracellular bacterial appendages with the PET molecules, wherein, the process of degradation begins. The bacteria employ certain enzymes like IsPETase and MHETase out of which the former hydrolyses PET into monomeric MHET (mono hydroxyethyl terephthalate) as a major part and dimeric BHET (Bis 2-hydroxyethyl terephthalate) as a minor part. The BHET molecules formed can further be degraded into monomeric MHET. Both of these intermediates are finally broken down into their safer monomeric forms viz. EG and TPA by the action of the latter enzyme MHETase (Fig. 2). These compounds, once formed, can be further biodegraded for the cell’s metabolic needs, purified and then can be recycled. The monomers formed are safe for the environment, unlike PET which can be highly inert and pose a serious threat to the environment if not recycled[15,18].
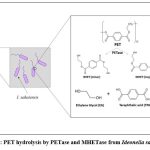 |
Figure 2: PET hydrolysis by PETase and MHETase from Ideonella sakaiensis |
Numerous microorganisms have been so far discovered and studied which could degrade different types of plastics (Table 2), I. sakaiensis being the most efficient of them for PET degradation.
Table 2: Depicts the list of different plastic degrading microorganisms along with their culture conditions and product yield.
|
S. No. |
Name of the microorganism |
Name of the enzyme |
Type of Plastic |
References |
Culture Conditions |
Product and yield |
|
1. |
Ideonella sakaiensis 201-F6 |
PETase |
PET |
10 |
Use of Consortium no. 46 incubated with PET film (diameter 6nm) at pH 7 and 30°C with change in MLE medium biweekly and YSV medium weekly. |
PET film was completely degraded after 6 weeks at 30°C. MHET was the major product released along with minor amounts of TPA and BHET. |
|
2. |
Fusarium solani |
FsC |
PET |
20 |
96 hr incubation with lcPET at 40°C |
5% film weight loss with only TPA after HPLC analysis |
|
3. |
Humicola insolens |
HiC |
PET |
20 |
96 hr incubation of lcPET at 70°C with 10 nmol/mL of HiC in 1 M Tris-HCl with 10% glycerol at pH 8 |
97∓ 3% film weight loss with only TPA after HPLC analysis |
|
4. |
Pseudomonas mendocina |
PmC |
PET |
20 |
96 hr incubation with lcPET at 50°C |
5% film weight loss with only TPA after HPLC analysis |
|
5. |
Streptomyces scabies |
Sub1 |
PET |
21 |
10 mg of PET granules incubated in 1 mL of Tris-HCl (20 mM, pH 7.5) with 3 μg of the enzyme Sub1 at 37 and 50°C for 20 days in presence of 0.5% Triton X-100. |
TPA was produced at the result of enzymatic hydrolysis with concentration of about 200μg/ml at 37°C and about 125 μg/ml at 50°C |
|
6. |
Clostridium botulinum |
Cbotu_EstA |
PET |
22,23 |
PET films (0.5 – 1 cm) incubated in 2mL 50mM citrate phosphate, pH 6.0 with 6mM Cbotu_EstA at 50°C for 11 days. |
Marginal concentration of Ta, ETa, ETaE was produced, however, genetic recombination with substitutions in Zn binding domain significantly improved it’s activity. |
|
7. |
Pseudomonas aeruginosa |
depolymerase |
PE |
24 |
1g PE beads incubated in liquid LCFBM medium with bacteria at 25 ◦C for 15 days at 180 rpm |
PE was degraded into PHAs with an average weight loss of 0.64% per day from total |
|
8. |
Phanerochaete chrysosporium, Trametes versicolor |
MnP |
PVC |
25 |
Inoculation of cellulose blended PVC films with 10ml fungal spores suspension (10 ± 2.1 × 106 spores mL -1) in MSM media (90ml) and incubated at 30°C for 3 months. |
CO2 (10.21g/L) was produced after 30 days, checked via FTIR. |
|
11. |
Thermobifida fusca |
Cutinase (TfCut2) |
PET |
26 |
PET nanoparticles incubated with 50 g mL−1 TfCut2 in 0.5 M Tris–HCl buffer at pH 8.5, 60 ◦C at 1000 rpm |
TPA, MHET & BHET were the main products with 0.55 g of MHET per gram PET and smaller amounts of TPA, BHET |
|
12. |
P. stutzeri |
PEG dehydrogenase |
PEG |
27 |
Incubation of the bacteria with 0.2% (wt vol-1) of 14,000 Da PEG for 30 hours |
100% degradation |
Enzymes involved in PET degradation
PET hydrolase (IsPETase)
Phylogenetic studies have revealed that this proteinaceous enzyme belongs to the family of cutinases and the class of esterases, i.e., enzymes which hydrolyze ester bonds into acid and alcohol via the help of water[2,18]. PETase was first discovered in Japan in 2016 in a bacterial strain (Ideonella sakaiensis strain 201-F6.) and is a principal PET degrading enzyme [2].
This extracellular enzyme is made up of three polypeptide chains viz. A, B and C chains [2,27]. Its active site is composed of a canonical α-/β-hydrolase fold with a conserved core consisting of seven α-helices and nine β-strands (a mix of parallel and antiparallel strands)[16,17]. The enzyme carries out hydrolysis with the help of a catalytic triad of Ser160-His237-Asp206 (including the signal peptide) (Fig. 3 (A)), out of which Serine acts as a nucleophile, polarized by histidine and in turn stabilised by the Aspartic acid [27,2].
According to some detailed crystallographic evaluations, it has been found that certain changes in the amino acid sequence (primary structure) of the enzyme aid in its catalytic properties significantly. For example, the presence of Trp156 above the active substrate-binding site provides a notable extent of flexibility, thereby enhancing its catalytic efficiency by significant folds. The catalytic triad also causes jiggling of Trp156, increasing pliability and making the binding of substrate molecules (PET) more convenient. The hydrophobic environment developed by Trp156 along with Tyr60 further aids in substrate binding. Another unique feature that distinguishes PETase from other cutinases is the presence of two disulfide bridges which contribute to the stability of the triad by holding close the His and Asp residues[18].
IsPETase differs from MHETase in the structure and composition of its active binding sites[27]. Thus, out of all PET hydrolytic enzymes (PHEs) discovered, PETase exhibits several times higher catalytic efficiency, making it a key fragment of plastic degradation[18].
MHET hydrolase (MHETase)
MHETase was first discovered in Japan in 2016 in the bacterial strain, Ideonella sakaiensis strain 201-F6. It belongs to the family of cutinases and the class of hydrolases[2]. The active site in MHET is covered by a special ‘lid domain’ which makes it unlike the other family members[18]. The presence of such a large lid domain aids in specific and accurate substrate binding with the enzyme[27]. The enzyme plays a crucial role in the process of PET degradation, after the hydrolytic breakdown of PET into MHET and TPA by the enzyme IsPETase, MHETase further acts on MHET and breaks it down into TPA and EG[28]. It shares a close association with the tannase/esterase family of enzymes, furthermore, it shares some structural similarities with its partner enzyme PETase[18].
Similar to IsPETase, the enzyme is also composed of an alpha/beta hydrolase fold with a core of catalytic triad composed of Ser225-His528-Asp492 residues (Fig. 3 (B)) flanked by a “lipase box”. However, instead of two, the core is edged by the presence of five disulphide linkages which contributes to the stability and compact arrangement of the catalytic triad. Unlike the IsPETase, this enzyme holds the substrate very tightly[2,24]. The significant changes in the working of MHETase from that of different catalysts occur because of loop insertions, cancellations, and recombinations[2].
 |
Figure 3: Structure of enzymes: PETase and MHETase |
Mechanism of PET degradation
PET degradation by I. sakaiensis is an enzymatic hydrolytic process undertaken by a set of two enzymes namely IsPETase (PET hydrolase) and MHETase (MHET hydrolase). I. sakaiensis, on coming in contact with the PET surface, binds with it firmly, which in turn triggers the release of the enzymes. It is believed that its extracellular appendages aid in the delivery of IsPETase (extracellular enzyme) on the PET surface, thus marking the beginning of the enzymatic degradation[16]. IsPETase is a hydrolytic enzyme which hydrolyses its substrate PET molecules via the involvement of its catalytic triad consisting of Ser-His-Asp residues. In the first step, the Ser molecule being the nucleophile, attacks the ester linkage on the acyl side, forming a covalent intermediate (Fig. 4) with the release of alcohol, which is ethylene glycol (EG) in this case[27]. The intermediates formed in this case are MHET (major) and BHET (minor). These products are transported in the periplasm for further actions with the help of porin proteins where MHETase is present as an outer membrane-anchored lipoprotein[16]. This tetrahedral intermediate formed, is stabilised by an oxyanion hole which is formed via covalent catalysis with Try87 and Met160 residues[18].
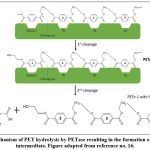 |
Figure 4: Mechanism of PET hydrolysis by PETase resulting in the formation of MHET as an intermediate. Figure adapted from reference no. 16. |
The second step involves the hydrolysis by MHETase which is also a hydrolytic enzyme, involved in the hydrolysis of MHET (major intermediate) to release the acid as well, which in this case is the TPA along with EG which are also its monomeric residues. TPA crosses the membrane into the cytosol (Fig. 5) where it is taken up by the TPA transporter along with TPA-binding protein and then integrated into the tricarboxylic acid (TCA) cycle via protocatechuic acid (PCA). The other product EG is metabolised in the TCA cycle via glyoxylic acid[27,16]. Thus, at the end of this two-step hydrolytic reaction, TPA and EG are formed as major products which can then be further processed to make new plastic polymers in plastic recycling plants.
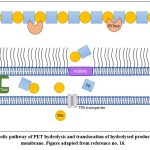 |
Figure 5: Metabolic pathway of PET hydrolysis and translocation of hydrolysed products across the cell membrane. Figure adapted from reference no. 16. |
A Viable Bio-Alternative to the Plastic Pollution Menace
Plastic is used widely all over the world for its durability, plasticity, transparency, convenient processing and low cost of production. It has a wide range of applications in markets like packaging, building and construction, automobiles, electrical equipment, and several household products; its demands rose even more especially during COVID when there was a surge in the requirement of plastic, as a significant number of medical equipments contain plastic. Apart from this, the use of masks, gloves and sanitizers increased several folds which added to the already existing heaps of plastics. The major concern with plastics is that it keeps accumulating in the environment and is highly resistant to enzymatic degradation. About 70 million tons of plastic has been produced in 2020 alone, leading to an enormous burden on the ecosystem[29,16,10].
Nevertheless, now it has a solution, the plastic-eating bacteria – Ideonella sakaiensis. This bacterium produces two types of enzymes viz. IsPETase and MHETase which can completely degrade PET type plastic and some other types as well. It utilises PET as a sole source of carbon for energy production. The mechanism of IsPETase action is unique, with stronger activity on PET films than other hydrolases/esterases and has the ability to use exceptionally big and hydrophobic polymers, according to the structural studies[30]. Its discovery has a promising implication towards recycling plastic waste. The process can be accelerated several folds with the assistance of the bacteria[31]. The enzymes can be extracted and then be further utilised on an industrial scale so as to recycle plastic. The traditional recycling methods use harmful chemicals whereas the use of bacterium provides a greener energy-saving alternative that on proper exploitation can fulfil the demand for sustainable waste management[32]. IsPETase shows higher activity on crystalline PET at lower temperatures and therefore can be an excellent option for marine microplastic degradation. The enzyme also has a potent role to play in the textile industry as PETase degrades PET fibres through eco-friendly hydrolysis and is therefore a great option over the conventional treatments which use strong alkaline NaOH solutions at high temperatures which not only deteriorates the environment but is an expensive cause of water pollution[33]. The active sites of I. sakaiensis are three in number, namely site 1, site 2 and site 3. These sites possess a specific binding energy with various types of plastic, the highest affinity being with polycarbonate and lowest being with polyvinyl chloride. The decreasing order of binding energy is as: polycarbonate > PET > polyamide > poly(methyl methacrylate) > polyurethane > polyvinyl chloride. High binding energy indicates higher energetic efficiency on hydrolysis by the enzyme[34].
Recent Advances and Future Prospects
In this world of ever-increasing plastic consumption, enzymatic degradation using Ideonella sakaiensis could prove to be a viable option for plastic recycling. Active research is ongoing to study the bacteria and its enzymes extensively, along with the development of novel techniques to derive benefit from its plastic degrading ability. Genetic engineering tools have been used to generate mutant forms of the enzyme IsPETase that show improvements in stability, catalytic efficiency and substrate specificity[33]. The wild type bacterial strain has undergone various genetic recombinations so far, with some of them yielding strains with much faster activity. One such recombination was carried out in the UK at the University of Portsmouth, wherein the functional expression of the enzymes IsPETase and MHETase was expressed in Chlamydomonas reinhardtii using a recombinant vector pBR9_PETase_Cre and in E. coli BL21 using vector pUCIDT plasmid (containing an indigenous signal peptide from I. sakaiensis) respectively. The enzymes were then recombined to produce a ‘super enzyme’ which could degrade PET plastic 6 times faster than the previously existing strains[2].
The flexibility of the protein is a key factor in determining the thermostability of the enzyme. Modification in the enzyme can enhance the thermostability, for instance, an introduction of disulfide bridges at suitable positions so that it does not impact the catalytic properties of the enzyme. Stefan et al. in their study discovered that Impranil agar plate screening shows an increase in the Tm of the wild type enzyme. IsPETaseTMK95N/F201I enzyme variant showed an improvement of 16.9°C. Replacing calcium-binding sites with disulfide bridges for Cut190 from Saccharomonospora viridis, a homologous enzyme of IsPETase, Tm could increase up to 20°C, leading to the discovery of N233C and S282C substitutions in IsPETase. Using N233C and S282C as a substitute in DuraPETase, contributed to increasing its thermostability. The highest Tm was found in the D1 variant of IsPETase, that is, 81.1°C[32]. Another study reported protein-based genetic engineering which soared up the active binding site of the enzyme IsPETase. During the experiment, two new variants (IsPETaseS242T and IsPETaseN246D) were genetically engineered from the wild strain (IsPETaseWT) which portrayed higher enzymatic activity. Likewise, a novel quadruple variant was developed by recombination of S242 T and N246D mutations with the variant IsPETaseS121E/D186H/R208A. The study was a big success as this quadruple variant manifested 58-fold increase in enzymatic efficiency by maintaining the degradation process for about 20 days, unlike the wild type strain which loses its activity in just about a day[35]. Though various advancements have already been made, many are still underway. Screening attempts to discover other novel PET degrading microorganisms from different habitats contaminated with plastics are ongoing.
Conclusion
Extensive use of plastics and poor recycling alternatives are leading to accumulation of millions of tons of plastics in terrestrials and marine environments. The recent discovery of microorganisms with plastic degrading abilities hold promise in bioremediation of this ever-growing plastic burden on Earth. PET degrading microbes are an interesting example of such useful microbes that have a potential in cleaning up the environment. The enzymes produced by these organisms have been a subject of extensive studies so that the mechanism of plastic degradation by them can be understood in more detail. Enzymatic degradation of PET is an advantageous process which cannot just be used to produce simpler TPA and MHET monomers but can also produce many high-value compounds via bio-conversion. Genetic engineering approaches are now being employed to improve them in terms of catalytic potential and stability. Though significant improvements have been made, still there is scope of more advancements in this field. For instance, plastic degradation may lead to the release of toxins in the environment, which on accumulation can have serious impacts. Plastic degradation also produces an impure mixture of its monomeric forms, therefore, recovery of desirable products from other elements adds on to the time and cost. Microbial degradation can although break down the non-biodegradable forms of plastic easily, still the idea of its scale-up remains a question as microbial culture, even today, isn’t cost-effective. Low thermal stability of IsPETase further remains a barrier in the commercialization of biodegradation. Effective strains of the bacteria are being genetically engineered along with the employment of different combinations of enzymes to combat these problems. With better techniques, reduced plastic production and public incentives, enzymatic plastic recycling, can serve as an excellent bioremediation tool to combat the problem of plastic pollution in near future.
Conflict of Interest
I hereby declare that all the authors and corresponds author do not have any conflict of interest.
Funding sources
There is not any external source of funding for conducting the research.
References
- Chen C. C., Han X., Ko T. P., Liu W., Guo R. T. Structural studies reveal the molecular mechanism of PETase. FEBS Journal. 2018;285(20):3717–3723.
CrossRef - Mathew J., Idicula Mathews A., Jose Vazhacharickal P. Microbial degradation of polyethylene terephthalate (PET): An outlook study. ~ 31 ~ Journal of Medicinal Plants Studies. 2021;9(5):31–40.
- Hiraga K., Taniguchi I., Yoshida S., Kimura Y., Oda K. Biodegradation of waste PET. EMBO Reports. 2019;20(11):e49365.
CrossRef - Jankauskaite V., Macijauskas G., & Lygaitis R. Polyethylene terephthalate waste recycling and application possibilities: a review. Mater Sci (Medžiagotyra). 2008;14(2):119-127
- Achilias D. S., & Karayannidis G. P. The chemical recycling of PET in the framework of sustainable development. Water, air and soil pollution: Focus. 2004;4(4):385-396.
CrossRef - Sinha V., Patel M. R., & Patel J. V. PET waste management by chemical recycling: a review. Journal of Polymers and the Environment. 2010;18(1):8-25.
CrossRef - Jeya G., Dhanalakshmi R., Anbarasu M., Vinitha V., & Sivamurugan V. A short review on latest developments in catalytic depolymerization of Poly (ethylene terephathalate) wastes. Journal of the Indian Chemical Society. 2022;99(1):100291.
CrossRef - Jeya G., Rajalakshmi S., Gayathri K. V., Priya P., Sakthivel P., & Sivamurugan V. A bird’s eye view on sustainable management solutions for non-degradable plastic wastes. In Organic Pollutants. 2022; pp 503-534.
CrossRef - Carta D., Cao G., & amp, D’Angeli C. Chemical recycling of poly (ethylene terephthalate)(PET) by hydrolysis and glycolysis. Environmental Science and Pollution Research. 2003;10(6):390-394.
CrossRef - Yoshida S., Hiraga K., Takehana T., Taniguchi I., Yamaji H., Maeda Y., Toyohara K., Miyamoto K., Kimura Y., Oda K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science. 2016;351(6278):1196–1199.
CrossRef - Yoshida S., Hiraga K., Taniguchi I., Oda K.: Ideonella sakaiensis, PETase, and MHETase: From identification of microbial PET degradation to enzyme characterization. In: Methods in Enzymology. 2021; pp 187–205.
CrossRef - Lee A., & Liew M. S. Tertiary recycling of plastics waste: an analysis of feedstock, chemical and biological degradation methods. Journal of Material Cycles and Waste Management. 2021;23(1):32-43.
CrossRef - Danso D., Schmeisser C., Chow J., Zimmermann W., Wei R., Leggewie C., Li X., Hazen T., Streit W. R. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Applied and Environmental Microbiology. 2018;84(8).
CrossRef - Tanasupawat S., Takehana T., Yoshida S., Hiraga K., Oda K. Ideonella sakaiensis sp. nov., isolated from a microbial consortium that degrades poly(ethylene terephthalate). International Journal of Systematic and Evolutionary Microbiology. 2016;66(8):2813–2818.
CrossRef - Widyastuti G. Genetic Engineered Ideonella sakaiensis Bacteria: A Solution of the Legendary Plastic Waste Problem. SSRN Electronic Journal. 2018.
CrossRef - Taniguchi I., Yoshida S., Hiraga K., Miyamoto K., Kimura Y., Oda K. Biodegradation of PET: Current Status and Application Aspects. ACS Catalysis. 2019;9(5):4089–4105.
CrossRef - Weiland M. H.: Enzymatic Biodegradation by Exploring the Rational Protein Engineering of the Polyethylene Terephthalate Hydrolyzing Enzyme PETase from Ideonella sakaiensis 201-F6. In: ACS Symposium Series. 2020; 1357: pp 161–174.
CrossRef - Maity W., Maity S., Bera S., Roy A. Emerging Roles of PETase and MHETase in the Biodegradation of Plastic Wastes. Applied Biochemistry and Biotechnology. 2021;193(8):2699–2716.
CrossRef - Ronkvist Å. M., Xie W., Lu W., Gross R. A. Cutinase-Catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules. 2009;42(14):5128–5138.
CrossRef - Jabloune R., Khalil M., ben Moussa I. E., Simao-Beaunoir A. M., Lerat S., Brzezinski R., Beaulieu C. Enzymatic Degradation of p-Nitrophenyl Esters, Polyethylene Terephthalate, Cutin, and Suberin by Sub1, a Suberinase Encoded by the Plant Pathogen Streptomyces scabies. Microbes Environ. 2020;35(1).
CrossRef - Perz V., Baumschlager A., Bleymaier K., Zitzenbacher S., Hromic A., Steinkellner G., … & Guebitz G. M. Hydrolysis of synthetic polyesters by Clostridium botulinum esterases. Biotechnology and bioengineering. 2016;113(5):1024-1034.
CrossRef - Biundo A., Reich J., Ribitsch D., & Guebitz G. M. Synergistic effect of mutagenesis and truncation to improve a polyesterase from Clostridium botulinum for polyester hydrolysis. Scientific reports. 2018;8(1):1-7.
CrossRef - Lee H. M., Kim H. R., Jeon E., Yu H. C., Lee S., Li J., & Kim D. H. Evaluation of the biodegradation efficiency of four various types of plastics by Pseudomonas aeruginosa isolated from the gut extract of superworms. Microorganisms. 2020;8(9):1341.
CrossRef - Kale S. K., Deshmukh A. G., Dudhare M. S., Patil V. B. Microbial degradation of plastic: a review. Journal of Biochemical Technology. 2015;6(2):952-961.
- Barth M., Oeser T., Wei R., Then J., Schmidt J., & Zimmermann W. Effect of hydrolysis products on the enzymatic degradation of polyethylene terephthalate nanoparticles by a polyester hydrolase from Thermobifida fusca. Biochemical engineering journal. 2015;93:222-228.
CrossRef - Wilkes R. A., Aristilde L. Degradation and metabolism of synthetic plastics and associated products by Pseudomonas sp.: capabilities and challenges. Journal of Applied Microbiology. 2017;123(3):582–593.
CrossRef - Graf L. G., Michels E. A. P., Yew Y., Liu W., Palm G. J., Weber G. Structural analysis of PET-degrading enzymes PETase and MHETase from Ideonella sakaiensis. In: Methods in Enzymology. 2021;648: pp 337–356.
CrossRef - Palm G. J., Reisky L., Böttcher D., Müller H., Michels E. A. P., Walczak M. C., Berndt L., Weiss M. S., Bornscheuer U. T., Weber G. Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nature Communications. 2019;10(1).
CrossRef - Dai L., Qu Y., Hu Y., Min J., Yu X., Chen C. C., Huang J. W., Guo R. T. Catalytically inactive lytic polysaccharide monooxygenase PcAA14A enhances the enzyme-mediated hydrolysis of polyethylene terephthalate. International Journal of Biological Macromolecules. 2021;190:456–462.
CrossRef - Seo H., Kim S., Son H. F., Sagong H. Y., Joo S., Kim K. J. Production of extracellular PETase from Ideonella sakaiensis using sec-dependent signal peptides in E. coli. Biochemical and Biophysical Research Communications. 2019;508(1):250–255.
CrossRef - Bornscheuer U. T. Feeding on plastic. Science.2016;351(6278):1154–1155.
CrossRef - Brott S., Pfaff L., Schuricht J., Schwarz J. N., Böttcher D., Badenhorst C. P. S., Wei R., Bornscheuer U. T. Engineering and evaluation of thermostable IsPETase variants for PET degradation. Engineering in Life Sciences. 2022;22(3–4):192.
CrossRef - Kan Y., He L., Luo Y., Bao R. IsPETase Is a Novel Biocatalyst for Poly(ethylene terephthalate) (PET) Hydrolysis. ChemBioChem. 2021;22(10):1706–1716.
CrossRef - Duru C. E., Duru I. A., Enyoh C. E. In silico binding affinity analysis of microplastic compounds on PET hydrolase enzyme target of Ideonella sakaiensis. Bulletin of the National Research Centre. 2021;45(1):1–8.
CrossRef - Son H. F., Joo S., Seo H., Sagong H. Y., Lee S. H., Hong H., Kim K. J. Structural bioinformatics-based protein engineering of thermo-stable PETase from Ideonella sakaiensis. Enzyme and Microbial Technology. 2020;14:109656.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





