Manuscript accepted on : 06-09-2023
Published online on: 04-10-2023
Plagiarism Check: Yes
Reviewed by: Dr. C. P. Sreena
Second Review by: Dr. Raghavendra Kumar Gunda
Final Approval by: Dr. Eugene A. Silow
P. Praveen Reddy1 and V. UmaMaheswara Rao2*
and V. UmaMaheswara Rao2*
1Department of Microbiology, School of Allied Health Sciences, Malla Reddy University, Maisammaguda, Dulapally, Hyderabad – 500100, Telangana, India
2Department of Botany and Microbiology, Acharya Nagarjuana University, Nagarjunanagar- 522510, Guntur Dt. Andhra Pradesh, India
Corresponding Author E-mail: umrvanga@yahoo.co.in
DOI : http://dx.doi.org/10.13005/bbra/3144
ABSTRACT: Human beings are heavily dependent on fossil fuels like coal and petroleum products for various daily activities in life. The large-scale usage of petroleum products releases different types of hazardous gasses, sulfur dioxide being one of them. The oxidation of organosulfur compounds in fuels release sulfur dioxide which is deleterious to humans and one of the causative factors for acid rains. The hydrodesulfurization, a conventional process is practiced for the elimination of sulfur from petroleum products during refining is not up to the mark for the total removal of sulfur content. Especially, highly recalcitrant organosulfur compounds like dibenzothiophene and its derivatives are more resistant to hydrodesulfurization. The biodesulfurization process which involves microorganisms for the removal of sulfur from petroleum products was suggested to be as the better alternative approach to hydrodesulfurization. It has been considered that dibenzothiophene as a reference model recalcitrant compound for biodesulfurization experiments and the microorganisms that exhibit 4S metabolic pathway for the elimination ofsulfur atom from dibenzothiophene as the potent desulfurizing strains. The 4S pathway is under the regulation of three genes (dszA, B and C) of dsz operon and they express the enzymatic proteins DszA(dibenzothiophene sulfone monooxygenase), DszB (hydroxyphenylbenzene sulfinate desulfinase) and DszC (dibenzothiophene monooxygenase), respectively. In the present study, the dszC gene pertaining to Streptomyces sp. VUR PPR 102 was made to produce corresponding sequence of DszC enzyme in National Centre for Biotechnology Information (NCBI) open reading frame finder. The amino acid sequence of DszC enzymatic protein was used in SWISS MODEL server and the three-dimensional model of DszC enzymatic protein was developed. The DszC model was validated in Rampage server, Swiss PDB Viewer, Verify3D and ERRAT servers.
KEYWORDS: ERRAT; DszC enzyme model; dszCgene; NCBI; PDB Viewer Verify 3D; Rampage; SWISS MODEL
Download this article as:| Copy the following to cite this article: Reddy P. P, Rao V. U. In Silico dszC Gene Analysis, Modeling and Validation of Dibenzothiophene monooxygenase (DszC Enzyme) of Dibenzothiophene Desulfurizing Streptomyces sp.VUR PPR 102. Biosci Biotech Res Asia 2023;20(3). |
| Copy the following to cite this URL: Reddy P. P, Rao V. U. In Silico dszC Gene Analysis, Modeling and Validation of Dibenzothiophene monooxygenase (DszC Enzyme) of Dibenzothiophene Desulfurizing Streptomyces sp.VUR PPR 102. Biosci Biotech Res Asia 2023;20(3). Available from: https://bit.ly/3Q1lUTF |
Introduction
In the present scenario, most of the daily activities and needs of human beings are associated with utilization of fossil fuels like petroleum products and coal. The petroleum products are continuously oxidized by automobiles and industries releasing hazardous gases. Sulfur dioxide is one such hazardous gas which is deleterious to humans and environment1. When sulfur dioxide concentration in atmosphere reaches to above 20 ppm concentration, it causes cough, irritation of eyes, alteration in heart-beat rhythm, secretion of mucus, chronic bronchitis, provocation of asthma and damage of nerves associated with respiratory system2.Sulfur dioxide in the environment is converted to sulfur trioxide (a secondary air pollutant) which in turn reacts with moisture to form sulfurous acid (further becomes sulfuric acid). The resulted acid combines with rain water and reaches the soil as acid rain. The acid rains increase the acidity in water bodies and affects aquatic life. In addition, acid rains decrease the pH of the soil drastically affecting the vegetation which results in the low yield of crop plants3.
The fossil fuels contain organosulfur compounds in the form of thiophenes, thiols and sulfides. When fossil fuels are burnt for their utilization, the sulfur dioxide is released due to the oxidation of these organosulfur compounds. The major organosulfur group responsible for sulfur dioxide emission is thiophene. Among the thiophenes, dibenzothiophene (DBT) is the most recalcitrant and abundantly present orgnaosulfur compound in fossil fuels4 and so considered as a model substrate for performing desulfurization studies5. Hydrodesulfurization (HDS), a contemporary method used for the removal of sulfur from petroleum products during refining process is not so efficient in fulfilling its task. Mainly, DBT and its alkyl derivatives are not affected and removed during HDS treatment. Biodesulfurization (BDS) process that exploits microbes to separate sulfur content from petroleum products was found and suggested as a better alternative approach to hydrodesulfurization. Moreover, BDS method is an economical and environmental friendly process. The microbes which can desulfurize DBT via 4S pathway are commercially important when compared with the microbes which metabolize DBT by other metabolic pathways which results in destruction of DBT ring structure and reduction of calorific value (combustion value) of the fuel (petroleum product).The microorganisms remove the sulfur atom via 4s metabolic pathway from DBT by breaking C-S (carbon-sulfur) bond without interrupting the cyclic ring structure of DBT and so, the mileage (calorific value) of fuel is not changed6. The four reaction steps of 4S pathway are catalyzed by DszA, B and C enzymes. The dszA, B and C genes (dsz operon) encode DszA, DszB and DszC enzymatic proteins, respectively. The enzyme, DszC (DBT monooxygenase) catalyze the first two consecutive reaction steps viz., conversion of DBT to DBTO (DBT oxide) and then DBTO to DBTO2 (DBT sulfone). In the third step, DBT sulfone is converted to HPBS (hydroxyl phenyl benzene sulfonate) by the activity of DszA enzyme (DBTO2 monooxygenase). The DszB enzyme (HPBS desulfinase) catalyze the conversion of HPBS to 2-HBP and sulfite, the final step of 4Spathway7.
Bioinformatics is a branch of science that deals with the use of software tools and programs in managing and interpretation of biological data. The bioinformatics tools can be employed for the In silico analysis of genes (in NCBI open reading frame finder), designing and modeling of biomolecules and evaluation of predicted models of biomolecules. Presently, protein modeling is an emerging area in the field of bioinformatics where in, various tools and programs are available for the prediction (SWISS MODEL server, Phyre2, NCBI blast etc.,) and validation
(Rampage server, PROCHECK server, ERRAT, ProSA etc.,) of three-dimensional structures ofproteins8,9,10.
The present work is designed to derive the protein sequence the dszC gene of Streptomyces sp. VUR PPR 102, isolated from oil contaminated soils11 , in open reading frame (ORF) finder of NCBI to obtain the sequence of DszC enzymatic protein and to predict its three-dimensional model by using SWISS MODEL server. Further, the three-dimensional model of DszC enzyme was checked for its validity in Rampage, SPBDV, Verify3D and ERRAT servers.
Materials and Methods
In Silico derivation of amino acid sequence of DszC enzyme from dszC gene
The dszC gene sequence of DBT desulfurizing Streptomyces sp. VUR PPR 102 was entered in open reading frame finder of NCBI in FASTA format. In NCBI ORF finder, the ORF tool generates an open reading frame (ORF) corresponding to the submitted gene sequence. The ORF is equivalent to a matured messenger RNA. The ORF contains the genetic code information for the sequence of amino acids of a protein to be synthesized. The ORF which is exhibiting maximum length is selected to generate corresponding sequence of a protein12,13.
Prediction of DszC enzyme model
The DszC enzyme sequence obtained was entered in SWISS MODEL server in FASTA format to built 3-D model of DszC enzyme protein. The DszC model was developed by automated mode. In SWISS MODEL server the submitted protein sequence protein (query protein sequence) is searched against various sequences of proteins in STML (template library). The protein of template library showing high degree of similarity to query protein sequence is regarded as template protein based on which the three-dimensional structure of query protein was predicted. The homology modeling of a protein in SWISS MODEL is accomplished by PROMOD3 modeling platform. The PROMOD3 employs HH search and BLAST for tracing the template sequences against the given protein sequence14,15.
Evaluation of DszC enzyme model quality
The DszC enzyme protein model was checked in Rampage, SPDBV, Verify3D and ERRAT. For checking the DszC enzyme protein model quality, PDB format of enzyme model was used. In Rampage server, Ramachandran plot was generated for the submitted protein model. The Ramachandran plot is produced based on Psi and Phi (torsion) angles of amino acid residues in the given protein model. Relying on these angles in the Ramachandran plot, amino acid distribution in favored, allowed and outlier regions is displayed. Depending upon the percentage of amino acids displayed in favored region, the quality of protein model was determined16,17. The PDB formats of DszC enzymatic protein and its template models were imported to SPDBV. Then, DszC enzyme model was superimposed on its template model to calculate root mean square deviation (RMSD) value. Based on the RMSD value the quality of the protein model was verified18. In verify3D, the consonance of protein model structure (3-D) with its amino acid sequence (1-D i.e., primary structure) is determined. The verify3D program predicts the better model quality of a protein structure19. The ERRAT program generates a plot which depicts the data of structural error of each amino acid in three-dimensional protein model. Then the overall quality factor is determined based on which model quality of protein is predicted20.
Results and Discussion
DszC enzyme protein sequence
In ORF finder of NCBI, for the submitted dszC gene sequence (Figure 1), six reading frames (ORFs) were obtained in the form of graphical bars (Figure 2). The actual reading frame length in each bar was indicated as shaded region. The reading frame starts with an initiation codon and terminates with a stop or nonsense codon. The reading frame with highest length was selected and translated to obtain the sequence of DszC enzyme21. The following was the amino acid sequence of DszC enzyme in FASTA format.
 |
Figure 1: Submission of dszC gene sequence in NCBI ORF finder
|
 |
Figure 2: Six open reading frames generated in ORF finder of NCBI
|
Homology modeling of DszC enzyme
To generate a model of protein in SWISS MODEL, its template library should contain at least one experimentally validated protein model which has a close resemblance to the submitted protein22. The protein sequence present in template library which was maximum similar to DszC enzyme sequence was treated as template and the template determined in SWISS MODEL was4doy.1.A. The alignment of sequences of template (4doy.1.A) and DszC enzyme was shown in Figure3. The three-dimensional structure of DszC enzyme (homodimer) (Figure4) was developed using the template, 4doy.1.A model (Figure5).
The DszC enzyme occurs as a tetramer which is formed by the combination of two homodimers. The monomers in each homodimer are intimately associated23. In the present work, in SWISS MODEL server a homodimer structure of DszC enzyme model is obtained. Hence, in the figure3 the alignment of two monomers (Model_1: A and Model_01: B) of DszC enzyme dimer with the template sequence was shown.
 |
Figure 3: Alignment of sequences of DszC enzymatic protein and its template in SWISS MODEL
|
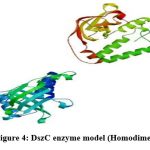 |
Figure 4: DszC enzyme model (Homodimer)
|
 |
Figure 5: Template (4doy.1.A)model
|
Verification of DszC enzyme model quality
The DszC enzyme model was evaluated by Ramachandran plot of Rampage server. The percentage (number) of amino acid residues of DszC enzyme model in favored, allowed and outlier regions (Table 1) confirm the validity of the model. In the Ramachandran plot (Figure 6) of DszC enzyme model 92.0% of amino acids were distributed in favored regions. In a Ramachadran Plot of a protein model with good stereo chemical quality, above 90% of amino acid residues will be in favored region24,25. In SPDBV, root-mean-square deviation (RMSD) value was obtained as a result of superimposition of DszC enzyme model over its template (Figure 7). The RMSD value infers the degree of similarity between two 3-D structures. The RMSD value generated between models of DszC enzymatic protein and its template was very low (0.26 Ao). The generated low RMSD value inferred a close similarity between the models, DszC enzymatic protein and its template which indicated the good quality of DszC enzyme model26,27. The verify3D program revealed that 100% of amino acid residues of DszC enzyme model had shown a 3D/1D profile score ≥ 0.2 (Figure 8) which indicated the best model quality of DszC enzyme. To confirm the validity of a protein model in Verify3D program, a minimum of 80% of its amino acids 3D/1D profile score must be ≥ 0.228. The ERRAT program computes overall quality factor for non-bonded linkages within the protein model. The highest quality factor scores infer the better quality of a protein. In general, the range of overall quality factor that can be accepted is > 50 for good protein model quality. The overall quality factor generated in ERRAT for DszC enzyme model was 91.765 (Figure 9) which indicated its genuine model quality20.
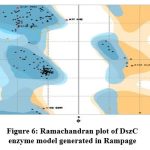 |
Figure 6: Ramachandran plot of DszC enzyme model generated in Rampage
|
Table 1: The abundance of amino acids of DszC enzyme model in different regions of Ramachandran plot of Streptomyces sp.VUR PPR 102
|
Amino acid residues in favored region (%) |
Amino acid residues in allowed region(%) |
Amino residues in outlier region(%) |
|
92.0% |
5.1% |
2.8% |
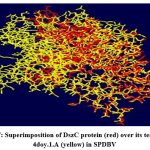 |
Figure 7: Superimposition of DszC protein (red) over its template, 4doy.1.A (yellow) in SPDBV
|
 |
Figure 8: Verify3D score for each amino acid residue in DszC enzyme model.
|
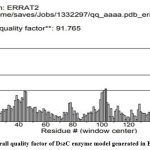 |
Figure 9: Overall quality factor of DszC enzyme model generated in ERRAT program
|
Conclusion
In the present work, the genomic and proteomic bioinformatics tools were exploited for the expression of dszC gene of DBT desulfurizing Streptomyces sp. VUR PPR 102 to produce the sequence of translated DszC enzymatic protein. The FASTA format sequence of DszC was entered in SWISS MODEL server to construct the three-dimensional model of DszC enzyme. The quality of modeled structure of DszC enzyme was evaluated using Rampage, SPDBV, Verify3D and ERRAT servers. Further, the DszC enzyme model can be modified to improve its catalytic activity by altering amino acids at some desired and specific sites using sophisticated software tools. The improved DszC enzyme activity can be checked by performing In Silico docking studies with its substrate (HPBS) using bioinformatics tools like Discovery Studio. The same improved DszC enzyme can be produced by microbes by making corresponding modifications in dszC gene and thus construction of genetically engineered microbes. Such microbes can be exploited in biodesulfurization of fossil fuels and thereby minimization of environmental pollution.
Acknowledgments
The authors express their thanks to Department of Botany and Microbiology, Acharya Nagarjuna University, Guntur, India for the cooperation and encouragement during the completion of present work. The first author extends thanks to the Management, Malla Reddy University, Hyderabad and Director and Dean, School of Allied Health Sciences, MRUH.
Conflict of Interest
The authors declare that they have no conflict of interest.
Funding sources
There are no funding sources.
References
- Bhanjadeo A.M, Rath K, Gupta D, Pradhan N, Biswal S.M, Mishra B.K and Subudhi U.Differential desulfurization of dibenzothiophene by newly identified MTCCstrains: Influence of operon array.PLoSOne.,2018;13(3):1-13.
CrossRef - Tunnicliffe W.S, Hilton M.F, Harrison R.M and Ayres J.G. The effect of sulfur dioxide exposure on indices of heart rate variability in normal and asthmatic adults. Eur Respir J.,2001;17(4):604-608.
CrossRef - Wondyfraw M. Mechanisms and effects of acid rain on environment. Journal of Earth Science & ClimaticChange.,2014;5(6):1-3.
- ConstantiM, Girait J and Bordons A. Desulfurization of dibenzothiophene by bacteria.World J Microb Biot., 1994;10:510-516.
CrossRef - Bordoloi N.K, Bhagowati P, Chaudhuri M.K and Mukherjee A.K. Proteomics and metaboliomics anaylyses to elucidate the desulfurization pathway of Chelatococcus sp. PLoSOne., 2016;11(4):1-21.
CrossRef - Labana S, Pandey G and Jain R.K. Desulfurization of dibenzothiophene and diesel oils by bacteria. Lett Appl Microbiol.,2005;40(3):159-163.
CrossRef - Mohebali G and Ball A.S. Biocatalytic desulfurization (BDS) of petrodiesel fuels. Microbiology.,2008;154:2169-2183.
CrossRef - Mehmood M.A, Sehar U and Ahamd N. Use of bioinformatics tools in different spheres of life sciences. J Data Mining Genomics Proteomics.,2014;5(2):1-13.
- Pruess M and Apweiler R. Bioinformatics resources for In Silico proteome analysis. J Biomed Biotechnol.,2003; 2003(4):231-236.
CrossRef - Fryer R.M, Randall J, Yoshida T, Hsiao L.L, Blumenstock J, Jensen K.E, Dimofte T,Jensen R.V and Gullans S.R. Global anaylsis of gene expression: Methods, interpretation, and pitfalls. Exp Nephrol.,2002;10(2):64-74.
CrossRef - Praveen R.P and UmaMaheswara R.V. Isolation and Identification of Dibenzothiophene Desulfurizing Bacteria Occurring in Oil Contaminated Soils of Mechanical Workshops. Jordan Journal of Biological Sciences., 2021; 14(4): 791-797.
CrossRef - Wheeler D.L, Church D.M, Federhen S, Lsh A.E, Madden T.L, Pontius J.U, Schuler G.D, Schriml L.M, Sequeira E, Tatusova T.A and Wagner L. Data base resources of the national center for biotechnology. NucleicAcidsRes.,2003;31(1):28-33.
CrossRef - Hung C.L and Lin C.Y. Open reading frame phylogenetic analysis on the cloud. Int J Genomics., 2013;2013:1-9.
CrossRef - Lam S.D, Das S, Sillitoea I and Orengo C. An overview of comparative modelling and resources dedicated to large-scale modeling of genome sequences. Acta Crystallographica D Struct Biol., 2017;73:628-640.
CrossRef - Arnold K, Bordoli L, Kopp J and Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics., 2006;22(2):195-201.
CrossRef - Chaudhary A, Goswami U and Singh H.S. Molecular characterization of Nippostrongylus brasiliensis (Nematode: Heligmosomatidae) from Musmusculus in India. Korean J Parasitol.,2016;54(6):743-750.
CrossRef - Satyanarayana S.D.V, Krishna M.S.R, Kumar P.P and Jeereddy S. In Silico structural homology modeling of nifA protein of rhizobial strains in selective legume plants. J Genet Eng Biotechnol., 2018;16(2),731-737.
CrossRef - Aslanzadeh V andGhaderian M. Homology Modeling and functional characterization ofPR-1a protein of Hordeum vulgare sub. sp. Vulgare. Bioinformation., 2012; 8(17): 807-811.
CrossRef - Kumar N, Tomar R, Pandey A, Tomar V, SinghV. K and Chandra R. Preclinical evaluation and molecular docking of 1,3-benzo dioxole propargyl ether derivatives as Novel inhibitor for combating the histone deacetylase enzyme in cancer. Artificial cells Nanomedicine and Biotechnology.,2018;46(6):1288-1299.
CrossRef - Singh S and Jha A. Structural characterization of 5-Hydroxytryptamine2A receptor in Homosapiens by In-Silico method. Asian J Pharm Clin Res.,2018;11(2):81-85.
CrossRef - Min X.L, Butler G, Storms R and Tsang A. Orf predictor: Predicting protein-codingregionsinEST-derived sequence. Nucleic Acids Res.,2005;33 (webserverissue), W677-W680.
CrossRef - Schwede T, Kopp J, Guex N and Peitsch M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res.,2003;31(13):3381-3385.
CrossRef - Guan L.J, Lee W.C, Wang S, Oshiro T, Izumi, Y, Ohtsuka J and Tanokura M. Crystal structures of apo-DszC and FMN-bound DszC from Rhodococcus erythropolisD-1.FEBS Journal.,2015;282(16):3126-3135.
CrossRef - Chandrasekaran G,H Wang E.C,Kang T.W, Kwon D.D, Park K, LeeJ.J and Lakshmanan V.K. Computational modeling of complete HOXB13 protein for predicting the functional effect of SNPs and the associated role in hereditary prostate cancer. Scientific Reports.,2017;7:1-18.
CrossRef - Beg M.A, Shivangi, Thakur S.C and Meena L.S. Structural prediction and mutational analysis of Rv39006c gene of Mycobacterium tuberculosis H37Rv to determine its essentiality in survival. Advances in Bioinformatics.,2018;2018:1-12.
CrossRef - Baloji G, Pasham S, Mahankali V, Garladinee M andAnkanagari S. Insights from the molecular docking analysis of phytohormone reveal brassinolide interaction with HSC70 from Pennisetum glaucum. Bioinformation.,2019;15(2):131-138.
CrossRef - Anupama P. In Silico studies on development of novel virostatic agents against bluetongue virus.ISRNVirology.,2014;2014:1-9.
CrossRef - Chikkerur J, Samanta A.K, Dhali A, Kolte A.P, Roy S and Maria P. In Silico evaluation and identification of fungi capable of producing endo-inulinase enzyme. PLoS One.,2018;13(7):1-19.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





