How to Cite | Publication History | PlumX Article Matrix
Beyond Silicon: The Advent of Biomolecular Computing
Department of Pharmacy, Sumandeep Vidyapeeth Deemed to be University, Piparia, Waghodia, Vadodara, Gujarat, India.
Corresponding Author E-mail: neil.dop@sumandeepvidyapeethdu.edu.in
DOI : http://dx.doi.org/10.13005/bbra/3169
ABSTRACT: Bio computing is an emerging interdisciplinary field that harnesses the information processing capabilities of biological substrates like DNA, proteins and cells to perform computational tasks. Rather than relying solely on conventional silicon-based computers, bio computing leverages the innate computational properties of biomolecules to encode, store, process and transmit information in unconventional ways. Core approaches include DNA computing, which uses DNA biochemistry to solve problems in a massively parallel fashion. Protein computing utilizes protein conformational dynamics to implement logic gates and communication modules for molecular information processing. Cellular computing focuses on engineering gene circuits and synthetic biology tools to program computational behaviours in living cells. Neural computing builds artificial neural networks inspired by biological brains. Key application areas include biomedicine, smart drug delivery systems, biosensing, hybrid organic-inorganic electronics, and biomolecular manufacturing. While still facing challenges around biocompatibility, programming complexity and ethical concerns, bio computing has achieved major technical milestones demonstrating its promise. Continued progress at the interface of biology and computing could enable future technologies like bio processors, in-vivo biocomputers, living materials and bio-intelligent systems. With responsible development, bio-inspired computation may catalyse the next revolution in human technological capabilities. This emerging field thus warrants enthusiastic attention as computation further converges with the living world.
KEYWORDS: Biocomputing; Bionics; Biologics; Bioelectronics; Wetware
Download this article as:| Copy the following to cite this article: Panchal N. B. Beyond Silicon: The Advent of Biomolecular Computing. Biotech Res Asia 2023;20(4). |
| Copy the following to cite this URL: Panchal N. B. Beyond Silicon: The Advent of Biomolecular Computing. Biotech Res Asia 2023;20(4). Available from: https://bit.ly/3MSCpiW |
Introduction
Bio computing is an exciting interdisciplinary field that utilizes biological materials to perform computations and information processing tasks.1 Instead of using traditional silicon-based computers, bio computing leverages the information storage and processing capabilities of biomolecules like DNA, proteins and cells. The key idea is that many biological molecules and systems already perform logic operations, data storage and communications as part of their normal functioning.2 Bio computing aims to understand these natural capabilities and engineer new synthetic biological systems to carry out useful computational tasks.3,4
Some examples of bio computing include
DNA computing – Using DNA and molecular biology tools to solve mathematical problems. DNA molecules can encode information and molecular operations on DNA like annealing can perform parallel computations.5,6
Protein computing – Using protein interactions and conformational changes to perform logic operations and calculations. Proteins can switch between different confirmations in response to inputs like other molecules binding, allowing them to mimic logic gates and circuits.7–10
Cellular computing – Programming gene circuits and networks within living cells to carry out sensing, information processing and actuation tasks. Synthetic biology allows engineering cells with toggle switches, oscillators, logic gates etc.11,12
Neural computing – Building artificial neural networks that are inspired by information processing in biological brains. The connections between neural network nodes mimic the synaptic signaling between neurons in the brain.13–16
Molecular computing – Designing and synthesizing molecules with specific structures so they can implement algorithmic functions and calculations when reacting with each other. The molecules effectively act as tiny programmable computers.17,18
Overall, bio computing provides an alternative paradigm to silicon computing by storing and processing information in biological substrates. It holds exciting promise for developing future biocompatible computing devices and interfacing them with biological systems.
History and Origins of Bio Computing
The foundations of bio computing were laid in the 1990s through pioneering work by computer scientists and biologists exploring the information processing capabilities of DNA and proteins.19,20 In 1994, Leonard Adleman demonstrated the first example of DNA computing by solving a small instance of the directed Hamilton Path problem using DNA molecules6,21. This seminal work established the possibility of using DNA biochemistry to perform computational operations.21
The field advanced significantly in subsequent years. In 1996, actual wet lab DNA computing systems were developed to solve chess problems and other computational challenges.22,23 Early proponents like Erik Winfree demonstrated simple DNA-based “robots” and computational circuits24,25. By 2002, gene regulatory networks were engineered to mimic neural network computations for pattern recognition tasks. 26–28
Protein computing also emerged in the 1990s, spearheaded by groups like Ehud Shapiro who designed in vitro enzymatic logic gates performing Boolean operations29–31. Other advances included designing molecular Turing machines based on proteins and using protein molecular recognition for biomolecular interfacing. The interdisciplinary field of synthetic biology greatly expanded the toolkit for engineering gene circuits and cellular computing systems.29,31,32
On the neural computing front, significant progress was made in modeling biological neurons and training artificial neural networks for pattern recognition and machine learning. Novel neural inspired algorithms like deep learning revolutionized fields like computer vision and natural language processing.33
Today, bio computing encompasses a diverse array of techniques harnessing DNA, proteins, cells, biomolecules and neural networks for information processing. Early visionary experiments have grown into a thriving research arena with conferences, journals and dedicated labs around the globe. Ongoing innovations promise an exciting future for biologically-inspired computation.34–36
Key Tools and Techniques Used in Bio Computing
Bio computing relies on the convergence of engineering and biotechnology to design, build, and optimize biological substrates for information processing and computation. This requires an extensive interdisciplinary toolkit to read, write, analyze and interface with DNA, proteins, cells and tissues.3
 |
Figure 1: Tools and Technologies of Bio-computing. |
DNA sequencing tools, such as next-generation sequencing, enable the rapid and cost-effective deciphering of genetic information within DNA, facilitating the design of synthetic gene circuits. Complementing this, directed evolution methods like error-prone Polymerase Chain Reaction (PCR) can be employed to engineer proteins and enzymes, tailoring them to exhibit specific computational properties. Meanwhile, rational protein engineering through techniques like site-directed mutagenesis refines protein structure and function, aligning them with precise computational requirements. These combined approaches empower researchers to craft custom biological components, paving the way for innovative applications in synthetic biology, biocomputing, and beyond.37–39
Gene synthesis techniques provide a cost-effective and rapid means to create novel genetic constructs for implementing biocomputational designs. Researchers can easily order synthetic genes from commercial vendors, receiving DNA fragments tailored to their specifications. This streamlined approach accelerates the development of customized biological components, facilitating the realization of innovative biocomputing applications across various fields, including synthetic biology and biotechnology.40 Automated liquid handling robotics enable efficient assembly of genetic circuits at high-throughput rates. Concurrently, CRISPR-Cas9 genome editing tools offer precise host cell genome modifications, optimizing computational designs by tailoring the cellular environment. This synergy of technologies accelerates the development of advanced bio-computational systems and applications, ranging from synthetic biology to biotechnology.41,42
Microfluidics technology revolutionizes bio-computing experiments by offering precise control over tiny fluid and cell volumes. Microfluidic chips, featuring integrated valves, channels, and chambers, enable the creation of programmable environments. Researchers harness this capability to conduct cellular computing and construct intricate biomolecular logic circuits. The versatility of microfluidics serves as a pivotal tool for advancing bio-computational research and applications in diverse fields, ranging from synthetic biology and biotechnology to cutting-edge biomedical engineering. Nanopore technology is a revolutionary approach to DNA/RNA sequencing, offering label-free single-molecule sensing. It operates through a nanoscale pore in a membrane. As DNA or RNA molecules are threaded through this pore, changes in electrical conductivity are detected in real-time, allowing for the precise identification of nucleotide sequences. This method eliminates the need for complex labeling procedures and provides high-resolution, rapid, and cost-effective sequencing. Nanopore technology holds great promise for genomics research, clinical diagnostics, and various applications requiring accurate and efficient molecular analysis.43,44
Fluorescence microscopy techniques, such as Fluorescence Resonance Energy Transfer (FRET) imaging, play a vital role in debugging genetic circuits by tracking molecular interactions within living cells. FRET relies on the principle that when two fluorophores are in close proximity, energy is transferred from one to the other, resulting in measurable fluorescence changes. By tagging molecules of interest with different fluorophores and observing their interactions through changes in fluorescence, researchers can gain insights into the behavior of genetic circuits in real-time, helping to optimize and debug their functionality.45 High-throughput screening tools are instrumental in testing extensive libraries of protein/DNA variants to identify those with the desired computational properties. The mechanism involves subjecting these variants to automated, rapid, and parallel assays, allowing the evaluation of their functional characteristics on a large scale. This screening process facilitates the selection of candidates that exhibit the most promising computational traits, expediting the development and optimization of biocomputational designs, such as synthetic gene circuits or protein-based computations.46 Biosensors are pivotal in biocomputing, serving as intermediaries that convert biological signals into measurable outputs and enable seamless interfacing with biocomputing systems. These devices typically consist of biological components, such as enzymes or antibodies, coupled with transducers that translate biological responses into electrical, optical, or other quantifiable signals. By detecting specific biomolecules or biological events, biosensors facilitate real-time monitoring, data acquisition, and signal processing within biocomputing systems, allowing for dynamic, responsive, and precise computational functions in various applications.47
Biocomputational modeling is instrumental in predicting and simulating the dynamics of gene circuits, protein interactions, and neuron behaviors before experimental implementation. Tools like the Systems Biology Markup Language (SBML) allow researchers to construct detailed mathematical models that represent biological system dynamics. These models incorporate parameters and equations to simulate the behavior of biological components, providing insights into how these systems function and respond to different inputs, ultimately aiding in the design and optimization of biocomputing systems and experiments.48–50
Ongoing advances in these core tools along with innovations in biomaterials, bioprinting, and bioelectronics promise to expand the capabilities of bio computing moving forward. The interdisciplinary toolkit combining engineering and biotechnology principles is key to realizing many of the futuristic applications envisioned in the field.51–53
Key Applications and Implementations of Bio Computing
Bio computing is catalyzing innovative applications across diverse domains including biomedicine, smart therapeutics, environmental sensing, materials science and hybrid bioelectronic devices. Researchers are harnessing the unique capabilities of biological substrates to design and engineer novel computational systems.
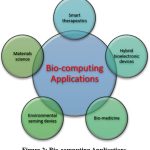 |
Figure 2: Bio-computing Applications |
In biomedicine, one prominent application area is developing systems for early disease diagnosis and continuous monitoring. Cancer detection systems have been demonstrated using artificial neural networks that analyze proteomic biomarkers in blood samples. By training on patient datasets, these Artificial Intelligence (AI) cancer classifiers can identify difficult to diagnose cancers like ovarian cancer based on biomarker profiles.54–56 Neural networks have also shown promise in medical image analysis, providing computer-aided diagnostics for improved treatment planning.57,58
Beyond diagnosis, bio computing is enabling smart drug delivery systems. Implantable bionanosensors have been proposed using protein logic gates to detect multiple biomarker inputs and decide molecular actuator functions accordingly. This biomolecular calculus, mimicking electronic circuits, allows intelligent therapeutic delivery tailored to personalized biomarker profiles.59–61 Portable smartphone-integrated biosensors are also being developed for rapid point-of-care diagnosis.62 Other efforts have explored engineering probiotic gut bacteria that sense pathogens in the gastrointestinal tract and secrete therapeutic compounds as desired.6364
In synthetic biology, engineered gene circuits and reprogrammed cells are being applied for portable and rapid disease screening. For instance, researchers designed whole-cell biosensors that produce a fluorescent output signal in response to the cancer biomarker interleukin-6.12,65 Such engineered living cells could offer continuous disease monitoring via implantable devices. Wearable fluorescent biosensors are also being integrated with smartphones for on-site diagnosis.66,67
Beyond medicine, DNA computing circuits have been applied for bio molecular analysis automation in Research and Development (R&D) labs. Systems have been engineered to solve optimization problems like the shortest path for analysing fluorescence microscopy images. DNA reaction networks that cascade over time act as chemical amplifiers for enhancing bioassay target detection. DNA tile self-assembly has also been leveraged to construct nucleic acid nanomaterials for drug delivery.68–70
Bio computing is also advancing hybrid bioelectronic systems, combining engineered biology with electronic interfaces. For example, Massachusetts Institute of Technology (MIT) designed an AI cancer classifier by using genetically engineered E. coli to target cancer biomarkers.71,72 The engineered bacteria provide molecular input data to train an electronic deep learning model for diagnostics. Such hybrids integrate the sensing/processing strengths of both biological and electronic substrates. 73
Environmental applications include engineering phage viruses that detect pollutant chemicals via colorimetric reactions and cells that luminesce in response to toxins.74 Plant nanobionics is creating “green computers” by embedding nanomaterials that monitor plant health. On the materials science front, viruses are being engineered to self-assemble into precise 2D and 3D structures for nanoscale fabrication.75,76
These examples highlight the diverse real-world promise as interdisciplinary bio computing transcends the lab bench. Ongoing advances in synthetic biology, AI and nanotechnology will further expand the application space and commercial potential.
Challenges and Limitations Currently Facing the Field of Bio Computing
One major challenge is creating biocompatible systems that can integrate and function effectively within biological environments and subjects. Biological tissues present a complex milieu of molecules, cell types and interactions that engineered systems must adapt to.77,78 Immunogenicity issues can arise whereby implanted bio computing devices trigger unwanted immune reactions. Approaches to improve biocompatibility include biomimetic designs using natural biological materials, bio-inert surface coatings, and localized release of immunosuppressant drugs.79–81
Programming and encoding complexity is another hurdle. Engineering robust gene circuits or neural networks requires sophisticated design tools, modeling frameworks, and debugging cycles.82 Synthetic biology is working to create modular, well-characterized genetic “parts” that can be predictably assembled. Abstraction layers and computer-aided design software also help hide low-level complexity. DNA sequence optimization algorithms assist in filling design specifications.83,84
Wet lab experimentation remains time-consuming and laborious. Standardizing protocols, automation technology like liquid handling robotics, and foundries for fabrication can relieve workflow bottlenecks.85,86 Microfluidics miniaturizes experiments onto chips and allows precise environmental control over reactions. High-throughput screening tools test libraries of design variants in parallel.87,88
Analysing and characterizing the dynamics of engineered networks is non-trivial. Researchers are devising mathematical models and multi-scale computational simulations to predict system behaviours before costly lab work.89 Advanced microscopy and “omics” tools facilitate quantitatively tracking molecular mechanisms.90
Maintaining the viability of engineered organisms and cells is an issue, as synthetic gene circuits add metabolic load. Strategies like genome streamlining, component optimization for low toxicity, and nutritional feedback controls help improve durability. Decoupling designs into separate survival and task-based modules also helps.11,12,77,91
Interfacing engineered systems with the complexity of real-world environments remains challenging. Bio-hybrid interfaces that connect synthetic biology with traditional electronics and hardware are still maturing. Onboard power sources or wireless power delivery are active research areas. Orthogonal communication schemes isolate synthetic systems from natural biological crosstalk.92,93
Safety and ethical concerns exist around bio computing applications like human augmentation or environmental release.94,95 Robust safeguards against unintended effects, molecular containment, and reversible engineering are important areas of investigation. Policy groups also advocate early awareness, monitoring and regulation around such engineering.
While significant hurdles exist, researchers are making steady progress through foundational engineering principles like modularity, model-based design, optimization, and characterization. Continued technology innovation and interdisciplinary collaboration will aid in systematically addressing the challenges on the path ahead.
Future Outlook and Emerging Trends in Bio Computing
Looking ahead, bio computing is poised to integrate more deeply with fields like artificial intelligence, robotics, and the Internet of Things. One avenue is developing bio-hybrid AI systems, combining biological computing substrates and learning algorithms for perception and inference tasks. Engineered organisms that synthesize their sensors and logic could enable fully autonomous, adaptable biocomputers.96–98
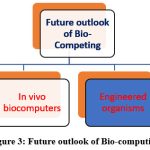 |
Figure 3: Future outlook of Bio-computing. |
Within the body, networks of engineered cells may one day run physiological regulation and repair routines like biological robots. In-vivo biocomputers could monitor organ health and coordinate therapeutic responses, forming a distributed treatment system. Nano-bioelectronics will miniaturize bio/organic interfaces for seamless integration. Implanted neural lace devices could allow direct brain-computer communication.12,91,99
DNA digital data storage is emerging as an ultra-dense, stable alternative to silicon memory. Entire datasets, books and videos have been encoded as DNA sequences. Integrating DNA memory with biological processors will enable storing and accessing massive information troves for AI. DNA could also allow on-chip training of nanoscale neural nets.100,101
Cell-free synthetic biology promises to expand bio manufacturing capabilities beyond living organisms. Printing hybrid bio-electronic materials containing engineered proteins and nucleic acids may support wearable, self-repairing soft robotics for human augmentation. Bio computing could thus distribute “enhanced intelligence” ubiquitously via engineered biomaterials.102,103
Self-organizing cellular systems that reshape and reconfigure on command will lead to programmable, morphing biohybrid materials for drug delivery or tissue engineering. Viral engineering for nanofabrication will create manufacturing platforms integrating top-down and bottom-up processes. Bio computing could thereby revolutionize digital fabrication, smart materials, and sustainable manufacturing.104–106
Protecting privacy and security of biometric data will be crucial as human-machine biointerfaces become intimate and pervasive. Ethical guidelines must shape applications for human improvement versus entrenching inequity. Overall, bio computing could fundamentally reshape our information infrastructure – while navigating immense opportunities and challenges along the way. Interdisciplinary collaboration, public awareness and appropriate regulations will help guide responsible progress.107,108
Ethical Considerations and Issues Surrounding Bio Computing
As with any powerful technology, bio computing carries risks of misuse along with immense potential for benefit. A major concern is the dual-use potential – capabilities meant for good could also be coopted for harmful use by malign actors. For instance, technologies for rapid vaccine development using synthetic biology could be misdirected towards engineering viral bioweapons. Strict biosafety measures and oversight are necessary to prevent misuse.109–112
Augmenting human abilities via neural implants or genetic engineering raises important ethical questions about human dignity, consent, and identity. Policy groups advocate for precautionary, step-wise integration of human enhancement technologies with ongoing ethical review. Access equality is also an issue – such technologies could widen social disparities if only accessible to the wealthy. 113–115
Applications like engineered viruses for nanofabrication or environmental remediation carry risks of unintended ecological impacts. Containment systems that prevent uncontrolled proliferation in the environment are critical. Tracking and recovery mechanisms should be incorporated as safeguards.116,117
Protecting the privacy of biological data like genomic profiles in biocomputing systems is imperative, given the sensitivity of such data. Techniques like data encryption, access controls and consent policies help secure users against violations of informational privacy.118
Broader societal impacts also need consideration. Emerging biotechnologies could disrupt economic sectors reliant on conventional manufacturing, agriculture, medicine etc. Policy foresight and planning will be essential to manage disruptions and harness benefits. Bio computing may only reach its full potential through open, inclusive public dialogue around hopes, concerns and what constitutes responsible development.119
Researchers are taking proactive steps to address ethical bio computing – calling for guidelines, risk assessments, red teams, codes of conduct, external oversight bodies and multidisciplinary perspectives. With diligent, ethical foundations guiding its progress, bio computing can usher in humanity’s next era of technological flourishing.120–122
Conclusion
In summary, this review has explored the emergence of bio computing, the interdisciplinary pursuit of harnessing biological substrates like DNA, proteins and cells for information processing and computation. We examined pioneering approaches like DNA computing, which leverages parallel molecular reactions to solve problems. Synthetic gene circuits and gene editing tools enable reprogramming cells into adaptive biocomputers. Protein engineering can construct molecular logic gates and communication circuits. Neural networks build on understandings of biological brains for machine learning.
These biomolecular and bio-inspired techniques showcase nature’s computational capabilities and offer alternatives to conventional silicon computers. However, significant challenges remain around biocompatibility, complexity, testing rigors and responsible development concerns. Prudent nurturing is vital to bridge the gap from tantalizing potential to real-world impact.
Looking forward, bio computing could transform application domains from biomedicine to smart materials, if key technical hurdles are overcome. This requires sustaining rigorous collaborative efforts across disciplines and emphasizing ethical stewardship. In summary, bio computing represents an auspicious convergence of biology and technology, unlocking new computational frontiers by interfacing silicon with carbon-based substrates. The decades ahead will prove pivotal in carefully charting this biology-technology frontier.
At its core, bio computing signals a conceptual fusion between the ancient programming inherent in life’s machinery and human engineering of biological systems. This field warrants enthusiastic nurturing to gently integrate engineered constructs within biological environments, opening new eras of flourishing. With diligent efforts, bio-inspired computation may someday emulate nature’s sophistication, ushering an organic computational age that advances humanity’s condition while stewarding all life.
Acknowledgement
We extend our heartfelt gratitude to Dr. A.K. Sheth and Dr. M.K. Mohan for their invaluable guidance and support during this research.
Conflicts of Interest
Our research and findings are driven solely by scientific merit and integrity, without any competing interests.
Funding Sources
The author has independently prepared this paper for publication based on their own research and efforts.
References
- Liang X, Zhu W, Lv Z, Zou Q. Molecular Computing and Bioinformatics. Molecules [Internet]. 2019 [cited 2023 Jul 26];24(13). Available from: /pmc/articles/PMC6651761/
- Grozinger L, Amos M, Gorochowski TE, Carbonell P, Oyarzún DA, Stoof R, et al. Pathways to cellular supremacy in biocomputing. Nat Commun 2019 101 [Internet]. 2019 Nov 20 [cited 2023 Jul 26];10(1):1–11. Available from: https://www.nature.com/articles/s41467-019-13232-z
- Goñi-Moreno A, Nikel PI. High-performance biocomputing in synthetic biology-integrated transcriptional and metabolic circuits. Front Bioeng Biotechnol. 2019 Mar 11;7(MAR):443035.
- Dixon TA, Williams TC, Pretorius IS. Sensing the future of bio-informational engineering. Nat Commun 2021 121 [Internet]. 2021 Jan 15 [cited 2023 Jul 26];12(1):1–12. Available from: https://www.nature.com/articles/s41467-020-20764-2
- Wang Z, Tan J, Huang D, Ren Y, Ji Z. A biological algorithm to solve the assignment problem based on DNA molecules computation. Appl Math Comput. 2014 Oct 1;244:183–90.
- Parker J. Computing with DNA. EMBO Rep [Internet]. 2003 Jan 1 [cited 2023 Jul 26];4(1):7. Available from: /pmc/articles/PMC1315819/
- Mhashal A, Emperador A, Orellana L. Computational techniques to study protein dynamics and conformations. Adv Protein Mol Struct Biol Methods. 2022 Jan 1;199–212.
- Walker JM. Molecular Modeling of Proteins, Methods in Molecular Biology. Methods [Internet]. 2015 [cited 2023 Jul 26];1215:390. Available from: http://link.springer.com/10.1007/978-1-59745-177-2
- Sannigrahi A, De N, Chattopadhyay K. The bright and dark sides of protein conformational switches and the unifying forces of infections. Commun Biol 2020 31 [Internet]. 2020 Jul 15 [cited 2023 Jul 26];3(1):1–6. Available from: https://www.nature.com/articles/s42003-020-1115-x
- Liu H, Dastidar SG, Lei H, Zhang W, Lee MC, Duan Y. Conformational changes in protein function. Methods Mol Biol [Internet]. 2008 [cited 2023 Jul 26];443:259–75. Available from: https://link.springer.com/protocol/10.1007/978-1-59745-177-2_14
- Sedlmayer F, Aubel D, Fussenegger M. Synthetic gene circuits for the detection, elimination and prevention of disease. Nat Biomed Eng 2018 26 [Internet]. 2018 Jun 11 [cited 2023 Jul 26];2(6):399–415. Available from: https://www.nature.com/articles/s41551-018-0215-0
- Cubillos-Ruiz A, Guo T, Sokolovska A, Miller PF, Collins JJ, Lu TK, et al. Engineering living therapeutics with synthetic biology. Nat Rev Drug Discov 2021 2012 [Internet]. 2021 Oct 6 [cited 2023 Jul 26];20(12):941–60. Available from: https://www.nature.com/articles/s41573-021-00285-3
- Kumarasinghe K, Kasabov N, Taylor D. Brain-inspired spiking neural networks for decoding and understanding muscle activity and kinematics from electroencephalography signals during hand movements. Sci Reports 2021 111 [Internet]. 2021 Jan 28 [cited 2023 Jul 26];11(1):1–15. Available from: https://www.nature.com/articles/s41598-021-81805-4
- Pircher T, Pircher B, Schlücker E, Feigenspan A. The structure dilemma in biological and artificial neural networks. Sci Reports 2021 111 [Internet]. 2021 Mar 10 [cited 2023 Jul 26];11(1):1–16. Available from: https://www.nature.com/articles/s41598-021-84813-6
- Kingsley G. Encyclopedia of Physical Science and Technology | ScienceDirect [Internet]. Academic Press. 2001 [cited 2023 Jul 26]. 133–143 p. Available from: https://www.sciencedirect.com/referencework/9780122274107/encyclopedia-of-physical-science-and-technology
- Strukov D. Building brain-inspired computing. Nat Commun 2019 101 [Internet]. 2019 Oct 18 [cited 2023 Jul 26];10(1):1–6. Available from: https://www.nature.com/articles/s41467-019-12521-x
- Kwon Y, Kang S, Choi YS, Kim I. Evolutionary design of molecules based on deep learning and a genetic algorithm. Sci Reports 2021 111 [Internet]. 2021 Aug 27 [cited 2023 Jul 26];11(1):1–11. Available from: https://www.nature.com/articles/s41598-021-96812-8
- Xu Y, Lin K, Wang S, Wang L, Cai C, Song C, et al. Deep learning for molecular generation. Future Med Chem [Internet]. 2019 Mar 1 [cited 2023 Jul 26];11(6):567–97. Available from: https://pubmed.ncbi.nlm.nih.gov/30698019/
- Hagen JB. The origins of bioinformatics. Nat Rev Genet 2000 13 [Internet]. 2000 [cited 2023 Jul 26];1(3):231–6. Available from: https://www.nature.com/articles/35042090
- Gauthier J, Vincent AT, Charette SJ, Derome N. A brief history of bioinformatics. Brief Bioinform [Internet]. 2019 Nov 1 [cited 2023 Jul 26];20(6):1981–96. Available from: https://pubmed.ncbi.nlm.nih.gov/30084940/
- Amos M. DNA Computing. Unconv Comput [Internet]. 2018 [cited 2023 Jul 26];307–25. Available from: https://link.springer.com/referenceworkentry/10.1007/978-1-4939-6883-1_131
- Faulhammer D, Cukras AR, Lipton RJ, Landweber LF. Molecular computation: RNA solutions to chess problems. Proc Natl Acad Sci U S A [Internet]. 2000 Feb 15 [cited 2023 Jul 26];97(4):1385–9. Available from: https://www.pnas.org/doi/abs/10.1073/pnas.97.4.1385
- Song T, Eshra A, Shah S, Bui H, Fu D, Yang M, et al. Fast and compact DNA logic circuits based on single-stranded gates using strand-displacing polymerase. Nat Nanotechnol 2019 1411 [Internet]. 2019 Sep 23 [cited 2023 Jul 26];14(11):1075–81. Available from: https://www.nature.com/articles/s41565-019-0544-5
- Reif JH. DNA robots sort as they walk. Science (80- ) [Internet]. 2017 Sep 15 [cited 2023 Aug 29];357(6356):1095–6. Available from: https://www.science.org/doi/10.1126/science.aao5125
- El-Seoud SA, Mohamed R, Ghoneimy S. DNA computing: Challenges and application. Int J Interact Mob Technol. 2017;11(2):74–87.
- Shu H, Zhou J, Lian Q, Li H, Zhao D, Zeng J, et al. Modeling gene regulatory networks using neural network architectures. Nat Comput Sci 2021 17 [Internet]. 2021 Jul 22 [cited 2023 Jul 26];1(7):491–501. Available from: https://www.nature.com/articles/s43588-021-00099-8
- Ho JWK, Charleston MA. Network modelling of gene regulation. Biophys Rev [Internet]. 2011 Mar 1 [cited 2023 Jul 26];3(1):1–13. Available from: https://link.springer.com/article/10.1007/s12551-010-0041-4
- Chen J, Wood DH. Computation with biomolecules. Proc Natl Acad Sci U S A. 2000 Feb 15;97(4):1328–30.
- Vishweshwaraiah YL, Chen J, Chirasani VR, Tabdanov ED, Dokholyan N V. Two-input protein logic gate for computation in living cells. Nat Commun 2021 121 [Internet]. 2021 Nov 16 [cited 2023 Jul 26];12(1):1–12. Available from: https://www.nature.com/articles/s41467-021-26937-x
- Shapiro E, Gil B. Biotechnology: logic goes in vitro. Nat Nanotechnol [Internet]. 2007 [cited 2023 Aug 29];2(2):84–5. Available from: https://pubmed.ncbi.nlm.nih.gov/18654224/
- Singh A. Designing protein logic gates. Nat Methods 2020 176 [Internet]. 2020 Jun 4 [cited 2023 Jul 26];17(6):565–565. Available from: https://www.nature.com/articles/s41592-020-0865-1
- Alfaro JA, Bohländer P, Dai M, Filius M, Howard CJ, van Kooten XF, et al. The emerging landscape of single-molecule protein sequencing technologies. Nat Methods 2021 186 [Internet]. 2021 Jun 7 [cited 2023 Jul 26];18(6):604–17. Available from: https://www.nature.com/articles/s41592-021-01143-1
- Woźniak S, Pantazi A, Bohnstingl T, Eleftheriou E. Deep learning incorporating biologically inspired neural dynamics and in-memory computing. Nat Mach Intell 2020 26 [Internet]. 2020 Jun 15 [cited 2023 Jul 26];2(6):325–36. Available from: https://www.nature.com/articles/s42256-020-0187-0
- Del Ser J, Osaba E, Molina D, Yang XS, Salcedo-Sanz S, Camacho D, et al. Bio-inspired computation: Where we stand and what’s next. Swarm Evol Comput. 2019 Aug 1;48:220–50.
- Miikkulainen R, Forrest S. A biological perspective on evolutionary computation. Nat Mach Intell 2021 31 [Internet]. 2021 Jan 18 [cited 2023 Jul 26];3(1):9–15. Available from: https://www.nature.com/articles/s42256-020-00278-8
- Xiong X, Zhu T, Zhu Y, Cao M, Xiao J, Li L, et al. Molecular convolutional neural networks with DNA regulatory circuits. Nat Mach Intell 2022 47 [Internet]. 2022 Jul 4 [cited 2023 Jul 26];4(7):625–35. Available from: https://www.nature.com/articles/s42256-022-00502-7
- Füllgrabe J, Gosal WS, Creed P, Liu S, Lumby CK, Morley DJ, et al. Simultaneous sequencing of genetic and epigenetic bases in DNA. Nat Biotechnol 2023 [Internet]. 2023 Feb 6 [cited 2023 Jul 26];1–8. Available from: https://www.nature.com/articles/s41587-022-01652-0
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol 2008 2610 [Internet]. 2008 Oct 9 [cited 2023 Jul 26];26(10):1135–45. Available from: https://www.nature.com/articles/nbt1486
- Meiser LC, Nguyen BH, Chen YJ, Nivala J, Strauss K, Ceze L, et al. Synthetic DNA applications in information technology. Nat Commun 2022 131 [Internet]. 2022 Jan 17 [cited 2023 Jul 26];13(1):1–13. Available from: https://www.nature.com/articles/s41467-021-27846-9
- Hughes RA, Ellington AD. Synthetic DNA Synthesis and Assembly: Putting the Synthetic in Synthetic Biology. Cold Spring Harb Perspect Biol [Internet]. 2017 Jan 1 [cited 2023 Jul 26];9(1). Available from: /pmc/articles/PMC5204324/
- Tegally H, San JE, Giandhari J, de Oliveira T. Unlocking the efficiency of genomics laboratories with robotic liquid-handling. BMC Genomics [Internet]. 2020 Dec 1 [cited 2023 Jul 26];21(1):1–15. Available from: https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-020-07137-1
- Ortiz L, Pavan M, McCarthy L, Timmons J, Densmore DM. Automated Robotic Liquid Handling Assembly of Modular DNA Devices. J Vis Exp [Internet]. 2017 Dec 1 [cited 2023 Jul 26];2017(130). Available from: https://pubmed.ncbi.nlm.nih.gov/29286379/
- Goto Y, Akahori R, Yanagi I, Takeda K ichi. Solid-state nanopores towards single-molecule DNA sequencing. J Hum Genet 2019 651 [Internet]. 2019 Aug 16 [cited 2023 Jul 26];65(1):69–77. Available from: https://www.nature.com/articles/s10038-019-0655-8
- Ding T, Yang J, Pan V, Zhao N, Lu Z, Ke Y, et al. DNA nanotechnology assisted nanopore-based analysis. Nucleic Acids Res [Internet]. 2020 Apr 6 [cited 2023 Jul 26];48(6):2791–806. Available from: https://pubmed.ncbi.nlm.nih.gov/32083656/
- Padilla-Parra S, Tramier M. FRET microscopy in the living cell: different approaches, strengths and weaknesses. Bioessays [Internet]. 2012 May [cited 2023 Jul 26];34(5):369–76. Available from: https://pubmed.ncbi.nlm.nih.gov/22415767/
- Liu Z, Chen S, Wu J. Advances in ultrahigh-throughput screening technologies for protein evolution. Trends Biotechnol. 2023 Apr 21;
- Mehrotra P. Biosensors and their applications – A review. J Oral Biol Craniofacial Res [Internet]. 2016 May 1 [cited 2023 Jul 26];6(2):153. Available from: /pmc/articles/PMC4862100/
- Wooley JC, Lin HS, Biology NRC (US) C on F at the I of C and. Computational Modeling and Simulation as Enablers for Biological Discovery. 2005 [cited 2023 Jul 26]; Available from: https://www.ncbi.nlm.nih.gov/books/NBK25466/
- Huang B, Jia D, Feng J, Levine H, Onuchic JN, Lu M. RACIPE: A computational tool for modeling gene regulatory circuits using randomization. BMC Syst Biol [Internet]. 2018 Jun 19 [cited 2023 Jul 26];12(1):1–12. Available from: https://bmcsystbiol.biomedcentral.com/articles/10.1186/s12918-018-0594-6
- Lopatkin AJ, Collins JJ. Predictive biology: modelling, understanding and harnessing microbial complexity. Nat Rev Microbiol 2020 189 [Internet]. 2020 May 29 [cited 2023 Jul 26];18(9):507–20. Available from: https://www.nature.com/articles/s41579-020-0372-5
- Dey M, Ozbolat IT. 3D bioprinting of cells, tissues and organs. Sci Reports 2020 101 [Internet]. 2020 Aug 18 [cited 2023 Jul 26];10(1):1–3. Available from: https://www.nature.com/articles/s41598-020-70086-y
- Agarwal S, Saha S, Balla VK, Pal A, Barui A, Bodhak S. Current Developments in 3D Bioprinting for Tissue and Organ Regeneration–A Review. Front Mech Eng. 2020 Oct 30;6:589171.
- Yilmaz B, Al Rashid A, Mou YA, Evis Z, Koç M. Bioprinting: A review of processes, materials and applications. Bioprinting. 2021 Aug 1;23:e00148.
- Elemento O, Leslie C, Lundin J, Tourassi G. Artificial intelligence in cancer research, diagnosis and therapy. Nat Rev Cancer 2021 2112 [Internet]. 2021 Sep 17 [cited 2023 Jul 26];21(12):747–52. Available from: https://www.nature.com/articles/s41568-021-00399-1
- Tran KA, Kondrashova O, Bradley A, Williams ED, Pearson J V., Waddell N. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med 2021 131 [Internet]. 2021 Sep 27 [cited 2023 Jul 26];13(1):1–17. Available from: https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-021-00968-x
- Shastry KA, Sanjay HA. Cancer diagnosis using artificial intelligence: a review. Artif Intell Rev [Internet]. 2022 Apr 1 [cited 2023 Jul 26];55(4):2641–73. Available from: https://link.springer.com/article/10.1007/s10462-021-10074-4
- Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK. Medical Image Analysis using Convolutional Neural Networks: A Review. J Med Syst. 2018 Nov 1;42(11).
- Jiang J, Trundle P, Ren J. Medical image analysis with artificial neural networks. Comput Med Imaging Graph [Internet]. 2010 Dec [cited 2023 Jul 26];34(8):617–31. Available from: https://pubmed.ncbi.nlm.nih.gov/20713305/
- Chang HJ, Zúñiga A, Conejero I, Voyvodic PL, Gracy J, Fajardo-Ruiz E, et al. Programmable receptors enable bacterial biosensors to detect pathological biomarkers in clinical samples. Nat Commun 2021 121 [Internet]. 2021 Sep 1 [cited 2023 Jul 26];12(1):1–12. Available from: https://www.nature.com/articles/s41467-021-25538-y
- Luo C, He L, Chen F, Fu T, Zhang P, Xiao Z, et al. Stimulus-responsive nanomaterials containing logic gates for biomedical applications. Cell Reports Phys Sci. 2021 Feb 24;2(2):100350.
- Amalfitano E, Pardee K. Logic invades cell-free biosensing. Nat Chem Biol 2022 184 [Internet]. 2022 Feb 17 [cited 2023 Jul 26];18(4):356–8. Available from: https://www.nature.com/articles/s41589-021-00963-8
- Qian S, Cui Y, Cai Z, Li L. Applications of smartphone-based colorimetric biosensors. Biosens Bioelectron X. 2022 Sep 1;11:100173.
- Hung YP, Lee CC, Lee JC, Tsai PJ, Hsueh PR, Ko WC. The Potential of Probiotics to Eradicate Gut Carriage of Pathogenic or Antimicrobial-Resistant Enterobacterales. Antibiotics [Internet]. 2021 Sep 1 [cited 2023 Jul 26];10(9). Available from: /pmc/articles/PMC8470257/
- Ma J, Lyu Y, Liu X, Jia X, Cui F, Wu X, et al. Engineered probiotics. Microb Cell Fact [Internet]. 2022 Dec 1 [cited 2023 Jul 26];21(1). Available from: /pmc/articles/PMC9044805/
- Zhao N, Song Y, Xie X, Zhu Z, Duan C, Nong C, et al. Synthetic biology-inspired cell engineering in diagnosis, treatment, and drug development. Signal Transduct Target Ther 2023 81 [Internet]. 2023 Mar 11 [cited 2023 Jul 26];8(1):1–21. Available from: https://www.nature.com/articles/s41392-023-01375-x
- Kim J, Campbell AS, de Ávila BEF, Wang J. Wearable biosensors for healthcare monitoring. Nat Biotechnol 2019 374 [Internet]. 2019 Feb 25 [cited 2023 Jul 26];37(4):389–406. Available from: https://www.nature.com/articles/s41587-019-0045-y
- Feiner R, Dvir T. Tissue–electronics interfaces: from implantable devices to engineered tissues. Nat Rev Mater 2017 31 [Internet]. 2017 Nov 28 [cited 2023 Jul 26];3(1):1–16. Available from: https://www.nature.com/articles/natrevmats201776
- Ma W, Zhan Y, Zhang Y, Mao C, Xie X, Lin Y. The biological applications of DNA nanomaterials: current challenges and future directions. Signal Transduct Target Ther 2021 61 [Internet]. 2021 Oct 8 [cited 2023 Jul 20];6(1):1–28. Available from: https://www.nature.com/articles/s41392-021-00727-9
- Ma Q, Zhang C, Zhang M, Han D, Tan W. DNA Computing: Principle, Construction, and Applications in Intelligent Diagnostics. Small Struct [Internet]. 2021 Nov 1 [cited 2023 Jul 26];2(11):2100051. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/sstr.202100051
- Li J, Green AA, Yan H, Fan C. Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat Chem 2017 911 [Internet]. 2017 Sep 25 [cited 2023 Jul 26];9(11):1056–67. Available from: https://www.nature.com/articles/nchem.2852
- Sharfstein ST. Bio-hybrid electronic and photonic devices. https://doi.org/101177/15353702221144087 [Internet]. 2022 Dec 19 [cited 2023 Jul 26];247(23):2128–41. Available from: https://journals.sagepub.com/doi/10.1177/15353702221144087
- Zhou N, Ma L. Smart bioelectronics and biomedical devices. Bio-Design Manuf [Internet]. 2022 Jan 1 [cited 2023 Jul 26];5(1):1–5. Available from: https://link.springer.com/article/10.1007/s42242-021-00179-8
- Riglar DT, Silver PA. Engineering bacteria for diagnostic and therapeutic applications. Nat Rev Microbiol 2018 164 [Internet]. 2018 Feb 5 [cited 2023 Jul 26];16(4):214–25. Available from: https://www.nature.com/articles/nrmicro.2017.172
- Favor AH, Llanos CD, Youngblut MD, Bardales JA. Optimizing bacteriophage engineering through an accelerated evolution platform. Sci Reports 2020 101 [Internet]. 2020 Aug 19 [cited 2023 Jul 26];10(1):1–10. Available from: https://www.nature.com/articles/s41598-020-70841-1
- Mony C, Kaur P, Rookes JE, Callahan DL, Eswaran S V., Yang W, et al. Nanomaterials for enhancing photosynthesis: interaction with plant photosystems and scope of nanobionics in agriculture. Environ Sci Nano [Internet]. 2022 Oct 13 [cited 2023 Jul 26];9(10):3659–83. Available from: https://pubs.rsc.org/en/content/articlehtml/2022/en/d2en00451h
- Giraldo JP, Landry MP, Faltermeier SM, McNicholas TP, Iverson NM, Boghossian AA, et al. Plant nanobionics approach to augment photosynthesis and biochemical sensing. Nat Mater 2014 134 [Internet]. 2014 Mar 16 [cited 2023 Jul 26];13(4):400–8. Available from: https://www.nature.com/articles/nmat3890
- Brooks SM, Alper HS. Applications, challenges, and needs for employing synthetic biology beyond the lab. Nat Commun 2021 121 [Internet]. 2021 Mar 2 [cited 2023 Jul 26];12(1):1–16. Available from: https://www.nature.com/articles/s41467-021-21740-0
- Low LA, Mummery C, Berridge BR, Austin CP, Tagle DA. Organs-on-chips: into the next decade. Nat Rev Drug Discov 2020 205 [Internet]. 2020 Sep 10 [cited 2023 Jul 26];20(5):345–61. Available from: https://www.nature.com/articles/s41573-020-0079-3
- Li Y, Li N, De Oliveira N, Wang S. Implantable bioelectronics toward long-term stability and sustainability. Matter. 2021 Apr 7;4(4):1125–41.
- Sunwoo SH, Ha KH, Lee S, Lu N, Kim DH. Wearable and Implantable Soft Bioelectronics: Device Designs and Material Strategies. Annu Rev Chem Biomol Eng [Internet]. 2021 Jun 7 [cited 2023 Jul 26];12:359–91. Available from: https://www.researchgate.net/publication/352200306_Wearable_and_Implantable_Soft_Bioelectronics_Device_Designs_and_Material_Strategies
- Petrus-Reurer S, Romano M, Howlett S, Jones JL, Lombardi G, Saeb-Parsy K. Immunological considerations and challenges for regenerative cellular therapies. Commun Biol 2021 41 [Internet]. 2021 Jun 25 [cited 2023 Jul 26];4(1):1–16. Available from: https://www.nature.com/articles/s42003-021-02237-4
- Gao B, Sun Q. Programming gene expression in multicellular organisms for physiology modulation through engineered bacteria. Nat Commun 2021 121 [Internet]. 2021 May 11 [cited 2023 Jul 26];12(1):1–8. Available from: https://www.nature.com/articles/s41467-021-22894-7
- Nandagopal N, Elowitz MB. Synthetic Biology: Integrated Gene Circuits. Science (80- ) [Internet]. 2011 Sep 2 [cited 2023 Jul 26];333(6047):1244–8. Available from: https://www.science.org/doi/10.1126/science.1207084
- Tang TC, An B, Huang Y, Vasikaran S, Wang Y, Jiang X, et al. Materials design by synthetic biology. Nat Rev Mater 2020 64 [Internet]. 2020 Dec 23 [cited 2023 Jul 26];6(4):332–50. Available from: https://www.nature.com/articles/s41578-020-00265-w
- Penders B, Horstman K, Vos R. Walking the Line between Lab and Computation: The “Moist” Zone. Bioscience [Internet]. 2008 Sep 1 [cited 2023 Jul 26];58(8):747–55. Available from: https://dx.doi.org/10.1641/B580811
- Ayoob JC, Kangas JD. 10 simple rules for teaching wet-lab experimentation to computational biology students, i.e., turning computer mice into lab rats. PLOS Comput Biol [Internet]. 2020 Jun 1 [cited 2023 Jul 26];16(6):e1007911. Available from: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1007911
- Paratore F, Bacheva V, Bercovici M, Kaigala G V. Reconfigurable microfluidics. Nat Rev Chem 2021 61 [Internet]. 2021 Dec 13 [cited 2023 Jul 26];6(1):70–80. Available from: https://www.nature.com/articles/s41570-021-00343-9
- Wang T, Yu C, Xie X. Microfluidics for Environmental Applications. Adv Biochem Eng Biotechnol [Internet]. 2022 [cited 2023 Jul 26];179:267–90. Available from: https://link.springer.com/chapter/10.1007/10_2020_128
- Freitas CGS, Aquino ALL, Ramos HS, Frery AC, Rosso OA. A detailed characterization of complex networks using Information Theory. Sci Reports 2019 91 [Internet]. 2019 Nov 13 [cited 2023 Jul 26];9(1):1–12. Available from: https://www.nature.com/articles/s41598-019-53167-5
- Kaur P, Singh A, Chana I. Computational Techniques and Tools for Omics Data Analysis: State-of-the-Art, Challenges, and Future Directions. Arch Comput Methods Eng 2021 287 [Internet]. 2021 Feb 1 [cited 2023 Jul 26];28(7):4595–631. Available from: https://link.springer.com/article/10.1007/s11831-021-09547-0
- Bashor CJ, Hilton IB, Bandukwala H, Smith DM, Veiseh O. Engineering the next generation of cell-based therapeutics. Nat Rev Drug Discov 2022 219 [Internet]. 2022 May 30 [cited 2023 Jul 26];21(9):655–75. Available from: https://www.nature.com/articles/s41573-022-00476-6
- Potts MW, Johnson A, Bullock S. Evaluating the complexity of engineered systems: A framework informed by a user case study. Syst Eng [Internet]. 2020 Nov 1 [cited 2023 Jul 26];23(6):707–23. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/sys.21558
- Sillitto H. Nature of an Engineered System: Illustrated from Engineering Artefacts and Complex Systems. Handb Syst Sci [Internet]. 2021 Jan 1 [cited 2023 Jul 26];983–1039. Available from: https://link.springer.com/referenceworkentry/10.1007/978-981-15-0720-5_17
- Beeckman DSA, Rüdelsheim P. Biosafety and Biosecurity in Containment: A Regulatory Overview. Front Bioeng Biotechnol. 2020 Jun 30;8:547059.
- Almeida M, Ranisch R. Beyond safety: mapping the ethical debate on heritable genome editing interventions. Humanit Soc Sci Commun 2022 91 [Internet]. 2022 Apr 20 [cited 2023 Jul 26];9(1):1–14. Available from: https://www.nature.com/articles/s41599-022-01147-y
- Bhardwaj A, Kishore S, Pandey DK. Artificial Intelligence in Biological Sciences. Life [Internet]. 2022 Sep 1 [cited 2023 Jul 26];12(9). Available from: /pmc/articles/PMC9505413/
- Moxley-Wyles B, Colling R, Verrill C. Artificial intelligence in pathology: an overview. Diagnostic Histopathol. 2020 Nov 1;26(11):513–20.
- Cui M, Zhang DY. Artificial intelligence and computational pathology. Lab Investig 2021 1014 [Internet]. 2021 Jan 16 [cited 2023 Jul 26];101(4):412–22. Available from: https://www.nature.com/articles/s41374-020-00514-0
- Meng X, Xing Y, Li J, Deng C, Li Y, Ren X, et al. Rebuilding the Vascular Network: In vivo and in vitro Approaches. Front Cell Dev Biol. 2021 Apr 21;9:639299.
- DNA storage is the most important innovation you’ve never heard about | TechRadar [Internet]. [cited 2023 Jul 26]. Available from: https://www.techradar.com/news/dna-data-storage-is-the-most-important-innovation-youve-never-heard-about
- Ceze L, Nivala J, Strauss K. Molecular Digital Data Storage using DNA. Nat Rev Genet [Internet]. 2019 May 8 [cited 2023 Jul 26]; Available from: https://www.microsoft.com/en-us/research/publication/molecular-digital-data-storage-using-dna/
- Lu Y. Cell-free synthetic biology: Engineering in an open world. Synth Syst Biotechnol. 2017 Mar 1;2(1):23–7.
- Kelwick RJR, Webb AJ, Freemont PS. Biological Materials: The Next Frontier for Cell-Free Synthetic Biology. Front Bioeng Biotechnol. 2020 May 12;8:541788.
- Brassard JA, Nikolaev M, Hübscher T, Hofer M, Lutolf MP. Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat Mater 2020 201 [Internet]. 2020 Sep 21 [cited 2023 Jul 26];20(1):22–9. Available from: https://www.nature.com/articles/s41563-020-00803-5
- Daly AC, Prendergast ME, Hughes AJ, Burdick JA. Bioprinting for the Biologist. Cell. 2021 Jan 7;184(1):18–32.
- Daly AC, Davidson MD, Burdick JA. 3D bioprinting of high cell-density heterogeneous tissue models through spheroid fusion within self-healing hydrogels. Nat Commun 2021 121 [Internet]. 2021 Feb 2 [cited 2023 Jul 26];12(1):1–13. Available from: https://www.nature.com/articles/s41467-021-21029-2
- Blanco-Gonzalo R, Lunerti C, Sanchez-Reillo R, Guest RM. Biometrics: Accessibility challenge or opportunity? PLoS One [Internet]. 2018 Mar 1 [cited 2023 Jul 26];13(3):e0194111. Available from: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0194111
- Jaswal G, Kanhangad V, Ramachandra R. AI and deep learning in biometric security : trends, potential, and challenges [Internet]. [cited 2023 Jul 26]. Available from: https://www.routledge.com/AI-and-Deep-Learning-in-Biometric-Security-Trends-Potential-and-Challenges/Jaswal-Kanhangad-Ramachandra/p/book/9780367422448
- Sandbrink JB, Koblentz GD. Biosecurity risks associated with vaccine platform technologies. Vaccine. 2022 Apr 14;40(17):2514–23.
- Zhou D, Song H, Wang J, Li Z, Xu S, Ji X, et al. Biosafety and biosecurity. J Biosaf Biosecurity. 2019 Mar 1;1(1):15–8.
- Langer R, Sharma S. The Blessing and Curse of Biotechnology: A Primer on Biosafety and Biosecurity – Carnegie Endowment for International Peace [Internet]. Vol. 9. 2020 [cited 2023 Jul 26]. p. 1–7. Available from: https://carnegieendowment.org/2020/11/20/blessing-and-curse-of-biotechnology-primer-on-biosafety-and-biosecurity-pub-83252
- DiEuliis D, Carter SR, Gronvall GK. Options for Synthetic DNA Order Screening, Revisited. mSphere [Internet]. 2017 Aug 30 [cited 2023 Nov 7];2(4). Available from: https://pubmed.ncbi.nlm.nih.gov/28861521/
- Cinel C, Valeriani D, Poli R. Neurotechnologies for human cognitive augmentation: Current state of the art and future prospects. Front Hum Neurosci. 2019 Feb 1;13:430907.
- Eden J, Bräcklein M, Ibáñez J, Barsakcioglu DY, Di Pino G, Farina D, et al. Principles of human movement augmentation and the challenges in making it a reality. Nat Commun 2022 131 [Internet]. 2022 Mar 15 [cited 2023 Jul 26];13(1):1–13. Available from: https://www.nature.com/articles/s41467-022-28725-7
- Almeida M, Diogo R. Human enhancementGenetic engineering and evolution. Evol Med Public Heal [Internet]. 2019 Jan 1 [cited 2023 Jul 26];2019(1):183–9. Available from: https://dx.doi.org/10.1093/emph/eoz026
- Linley S, Thomson NR. Environmental Applications of Nanotechnology: Nano-enabled Remediation Processes in Water, Soil and Air Treatment. Water, Air, Soil Pollut 2021 2322 [Internet]. 2021 Feb 5 [cited 2023 Jul 26];232(2):1–50. Available from: https://link.springer.com/article/10.1007/s11270-021-04985-9
- Bradford SA, Shen C, Kim H, Letcher RJ, Rinklebe J, Ok YS, et al. Environmental applications and risks of nanomaterials: An introduction to CREST publications during 2018–2021. https://doi.org/101080/1064338920212020425 [Internet]. 2021 [cited 2023 Jul 26];52(21):3753–62. Available from: https://www.tandfonline.com/doi/abs/10.1080/10643389.2021.2020425
- Bonomi L, Huang Y, Ohno-Machado L. Privacy Challenges and Research Opportunities for Genomic Data Sharing. Nat Genet [Internet]. 2020 Jul 1 [cited 2023 Jul 26];52(7):646. Available from: /pmc/articles/PMC7761157/
- Asveld L, Osseweijer P, Posada JA. Societal and ethical issues in industrial biotechnology. Adv Biochem Eng Biotechnol [Internet]. 2020 [cited 2023 Jul 25];173:121–41. Available from: https://link.springer.com/chapter/10.1007/10_2019_100
- Wolpe PR, Rommelfanger KS. Ethical principles for the use of human cellular biotechnologies. Nat Biotechnol 2017 3511 [Internet]. 2017 Nov 9 [cited 2023 Jul 26];35(11):1050–8. Available from: https://www.nature.com/articles/nbt.4007
- Nebeker C, Torous J, Bartlett Ellis RJ. Building the case for actionable ethics in digital health research supported by artificial intelligence. BMC Med [Internet]. 2019 Jul 17 [cited 2023 Jul 26];17(1):1–7. Available from: https://bmcmedicine.biomedcentral.com/articles/10.1186/s12916-019-1377-7
- Ferretti A, Ienca M, Sheehan M, Blasimme A, Dove ES, Farsides B, et al. Ethics review of big data research: What should stay and what should be reformed? BMC Med Ethics [Internet]. 2021 Dec 1 [cited 2023 Jul 23];22(1):1–13. Available from: https://bmcmedethics.biomedcentral.com/articles/10.1186/s12910-021-00616-4

This work is licensed under a Creative Commons Attribution 4.0 International License.






