How to Cite | Publication History | PlumX Article Matrix
Chlorpyrifos Degradation by Bacillus tropicus a Plant Growth Promoting Rhizobacteria
Deepak KumarMalik1* , Raina Singhmar1
, Raina Singhmar1 , Vivek Singh1
, Vivek Singh1 , Meenu Rathi2 and Vishal Ahlawat3
, Meenu Rathi2 and Vishal Ahlawat3
1Department of Biotechnology, University Institute of Engineering and Technology, Kurukshetra University, Kurukshetra, Haryana, India.
2Department of Botany, Gandhi Memorial National College, Ambala Cantt, Haryana, India.
3Department of Mechanical Engineering, University Institute of Engineering and Technology, Kurukshetra University, Kurukshetra, Haryana, India.
Corresponding Author E-mail: dmalik2015@kuk.ac.in
DOI : http://dx.doi.org/10.13005/bbra/3230
ABSTRACT: Excessive use of organophosphate in modern farming to improve the crop productivity has cause pollution in soil, water and air which lead environmental as well as human hazards. Microbial fertility is adversely affected by the use of pesticides. Thus, the present study focused on the isolation and screening of effective isolates with multi-traits PGPR activities and further studied for chlorpyrifos pesticide degradation. The bacterial isolate DK5 was showing multiple PGPR activity, identified as Bacillus tropicus by 16S rRNA sequencing. The chlorpyrifos degradation by isolated Bacillus tropicuswas studied by using resting cell study. In HPLC analysis revealed that Bacillus tropicus degrade 60% chlorpyrifos after 48 hrs. of incubation followed by 99% after 72 hrs. of incubation. Generally, results of this study revealed that isolate DK5 identified as Bacillus tropicus can be used for the successfully removal of chlorpyrifos from contaminated soil and plant growth promotion.
KEYWORDS: Chlorpyrifos; HPLC; Pesticide; PGPR
Download this article as:| Copy the following to cite this article: Malik D, K, Singhmar R, Singh S, Rathi M, Ahlawat V. Chlorpyrifos Degradation by Bacillus tropicus a Plant Growth Promoting Rhizobacteria. Biotech Res Asia 2024;21(1). |
| Copy the following to cite this URL: Malik D, K, Singhmar R, Singh S, Rathi M, Ahlawat V. Chlorpyrifos Degradation by Bacillus tropicus a Plant Growth Promoting Rhizobacteria. Biotech Res Asia 2024;21(1). Available from: https://bit.ly/3IT0y6A |
Introduction
Farmers in India are using excessive pesticides and chemical fertilisers to combat pests and infections.1 Pesticides such as Malathion, carbofuran, Diazinon, Methyl parathion, Endosulphan, Monostrophes and Chlorpyrifos are commonly used in agriculture crop. 2 The widespread application of pesticides contaminates the soil ecosystem and raise the health issue. 3 Pesticides alter soil microflora over long run, reducing soil fertility and agricultural yield. 4 Degradation of such pollutants is crucial in agricultural practise in order to minimise the pesticide loads from the soil and provide healthy food and a healthier environment for future generations. A reliable method of degrading the pesticides is called bioremediation. Phytohormone synthesis, mineral solubilization, N2-fixation and other plant growth-promoting activities are reported in pesticide degrading microorganisms.5 Bioremediation is a technique that provide a cost-effective and dependable way of pesticide elimination form the soil. Plant Growth Promoting Rhizobacteria (PGPR) have great promise for boosting plant biomass and minimising the phytotoxic effects of organic contaminants.6 PGPR having the ability to degrade the pesticides provides, clean environment without any use of chemical fertilizers.7 The application of PGPR for bioremediation of soil contaminated with heavy metals, crude oil and pesticides is very new area of research.8-10 In this study we are reporting the chlorpyrifos degrading bacterial stain with plant growth promoting activity.
Material and Methods
Materials
Commercial-gradeChlorpyrifos was purchased from the local market of Kurukshetra and reference material was purchased from Sigma-Aldrich. HPLC grade Hexane, Ethyl acetate, Acetonitrile and HPLC water were purchased from HI Media. The chemicals used were of analytical grade.
Isolation of pesticide degrading bacteria
The enrichment process was used for the screening and isolation of chlorpyrifos degrading bacteria. The soil sample was taken from the rice rhizosphere soil of village Kirmich, Kurukshetra, Haryana. Soil sample was transferred to Basal Salt Medium (BSM) containing 0.4mM chlorpyrifos and kept on a rotary shaker at 80 rpm and 35°C for 1 week. After one week of incubation, culture supernatant was again transferred in to the BSM medium with 0.45mM increasing concentration of chlorpyrifos. The final round of enrichment was carried out by using 0.50 mM increasing concentration of chlorpyrifos. The enrichment media was diluted and spread over nutrient agar plates for the segregation of bacterial strains. Bacterial colonies were purified by streaking-over nutrient agar plates. All rhizospheric colonies were screened for plant growth promoting activity.
Screening of isolated bacterial colonies for plant growth promoting activity
Phosphate solubilization, Potassium solubilization and zinc solubilization
The isolated bacterial colonies were spot inoculated on Pikovskaya’s medium plates, incubated at 28°C for 5–7 days to measure the phosphate solubilization activity zone.11 To observe the potassium solubilization zone, the isolated bacterial cultures were spot-inoculated on Aleksandrow Agar medium and incubated at 28°C for 3 to 7 days.12 To check the solubilization of zinc, bacterial isolates were spot-inoculated on modified agar plates containing 0.1% zinc carbonate and incubated at 28°C for 7 days.13
IAA, Ammonium production, HCN production and antibiotic susceptibility test
Isolated bacterial colonies were added to nutrient broth infused with 0.1% tryptophan followed by incubation at 28°C for 48 hr. Nutrient broth was centrifuged after incubation and culture supernatant was mixed with equal amount of Salawaski’s reagent which was incubated for 30 min at room temperature to observe the color change from yellow to pink or red. The quantitative production of IAA by bacterial isolate was measured at 530 nm.14 Spot inoculated plate was incubated for 4 days at 30°C with a Whatman filter paper placed inside the plate’s lid after being soaked with 0.5% picric acid solution. HCN synthesis was indicated by change in the light brown to dark brown color production.15 The isolated bacterial colonies were cultivated overnight in 10 mLof peptone broth kept at 35°C. Post incubation 0.5 nessler’s reagent was added to peptone broth, change of color from yellow to dark brown indicate the ammonia production.16 The antibiotic susceptibility was checked against different antibiotic by using disc diffusion method using on Nutrient agar plates.17
Chlorpyrifos degradation by bacterial isolate DK5
Bacterial isolate was tested for their ability to degrade chlorpyrifos by resting cell studies. The isolate was grown in 300 mL nutrient broth supplemented with 0.5 mM chlorpyrifos in shaking condition at 30°C up to mid log phase. Isolate harvested at 4°C, cells were washed twice with MM. These cells were resuspended in 50 mL BSM supplemented with 0.5 mM chlorpyrifos. Aliquots of 10 mLwere taken at different time interval of 0, 12, 24 and 48 hr. Chlorpyrifos was extracted from cell free supernatant by extraction with equal amount of Hexane and Ethyl acetate. Excess of organic solvent was evaporated by using rotavapor before the estimation of chlorpyrifos degradation using HPLC. 18 HPLC for detection of chlorpyrifosa mobile phase of Acetonitrile–ultrapure water (50:50 v/v) at a flow rate of 1.2 mL/min, 20 µL of the sample was injected at 30°Cand detected at 254 nm. The degradation percentages were calculated with respect to area (mAU.s).
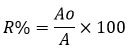
Where, R% is percentage of pesticide, Ao is area under extract and A is area under control peak of chlorpyriphos.
Results
Screening and Identification of PGPR
Five morphologically different bacterial strains were isolated by using enrichment method technique. Isolated bacterial isolates were coded as DK1 to DK5 and screened for the plant growth promoting activity. The potassium solubilisation was shown by bacterial isolate DK5 on Aleksandrow agar. The bacterial isolate DK5 was showing HCN, IAA and ammonium production with phosphate, Potassium, zinc solubilisation ability. Bacterial isolate DK5 was also showing good antibiotic susceptibility as compared to other isolates as shown in Table1.
Table 1: Plant growth promoting activity of isolated bacteria strains from contaminated soil.
| Isolates | Phosphate solubilization | Potassium solubilization | Zinc solubilization | IAA Production | Ammonium production | HCN Production | Antibiotic susceptibility |
| DK1 | – | – | – | + | + | + | – |
| DK2 | – | – | – | + | – | – | – |
| DK3 | – | – | – | + | – | – | – |
| DK4 | – | – | – | + | – | – | – |
| DK5 | + | + | + | + | + | + | + |
The zone of potassium solubilisation efficiency shown by bacterial isolates DK5 was measured as 17 mm. DK5 indicates its substantial capacity to release potassium from insoluble sources, for use as a biofertilizer or bioinoculant in sustainable agriculture. [17,18] On the basis of plant growth promoting activity and growth in MSM medium containing the 0.5mM chlorpyrifosbacterial isolate DK5 selected for further study. Bacterial isolate DK5 was characterized by 16S rRNA sequencing from NCIM, Pune, India. The DK5 16S RNA sequence was subjected to nBLAST analysis, which showed 99.72% similarity with Bacillus tropicus. Phylogenetic was constructed by using neighbour joining method for estimating the distance between evolutionary species as shown in Figure 1.
 |
Figure 1: Phylogenetic tree of bacterial isolate DK5 constructed using neighbour joining method. |
Quantitative analysis of chlorpyrifos degradation
To elucidate bacterial isolate DK5 degrades chlorpyrifos, resting cell study was performed. After HPLC analysis, 40% degradation at 24 hrs., 60% degradation at 48 hrs. and 100% degradation at 72 hrs. were found as compare to the initial concentration of chlorpyrifos as shown in Figure 2.
 |
Figure 2: HPLC chromatograph of chlorpyrifos pesticide degradation (a) zero degradation(b) 40% degradation rate after 24 hr (c) 60% degradation at 48 hr(d) at 72 hr the chlorpyrifos is completely degrade. |
Discussion
The present study isolated and identified a potential PGPR coded as DK5, that was able to solubilize potassium and produce HCN, IAA, and ammonium, as well as solubilize phosphate, potassium, and zinc. The ability to solubilize potassium is crucial for enhancing nutrient availability to plants, as potassium is an essential macronutrient that plays a vital role in various physiological processes within plants. The production of HCN and IAA further supports its role as a plant growth-promoting bacterium, as these compounds have been linked to increased plant growth, root development, and disease resistance. 19 The observation of good antibiotic susceptibility in DK5 is encouraging, as it implies that this strain may be safer and more viable for potential agricultural applications, given concerns regarding antibiotic resistance and its impact on environmental and human health. 20-21 In this study, DK5 was able to grow in MSM medium containing 0.5 mM chlorpyrifos, indicating its potential as a chlorpyrifos-degrading PGPR.
Resting cell studies were performed to elucidate DK5 ability to degrade chlorpyrifos, and it was found that DK5 was able to completely degrade pesticide after at 72 hours of incubation as compared to the initial concentration of chlorpyrifos. However, in case of heat killed resting cells no depletion of chlorpyrifos was observed at any time point. Several bacteria including Pseudomonas, Arthrobacter, Bacillus, Klebsiella, Ochrobactrum Flavobacterium and Agrobacterium have been isolated and found that are capable of degrading pesticides but do not necessarily support plant growth and can be pathogenic.22 Bacterial isolate Bacillus cereus CT3 isolated from cotton growing soil showed 88% degradation of chlorpyrifos in 8 days.23 In a similar study organophosphate pesticide degrading bacteria identified as Arthrobacter sp. HM01 was able to degrade chlorpyrifos and aided in plant growth promotion activity.24 These findings open up possibilities for using this bacterial strain in bioremediation efforts to address chlorpyrifos contamination, leading to potential advancements in environmental sustainability.
Conclusion
The bacterial isolate DK5, identified as Bacillus tropicus, exhibited the capability to efficiently degrade chlorpyrifos within a 72-hour timeframe, as confirmed by High-Performance Liquid Chromatography (HPLC) analysis. Bacillus tropicus DK5 was able to use chlorpyrifos as carbon source for plant growth promotion activity. The findings of this research offer promising prospects for the development of a new bio-inoculant in agriculture. By combining the ability to mitigate pesticide toxicity with plant growth promotion, DK5 holds the potential to enhance crop yield and contribute to sustainable agricultural practices. Further field trials can be performed to commercialize this as bio-fertilizer.
Acknowledgement
We gratefully acknowledge the use of Biochemistry Lab facilities at UIET, Kurukshetra University for providing necessary equipment’s, infrastructure and technical support that enabled us to conduct our experiments and analyze the data.
Declaration
I declare that the manuscript has not been published in any journal/book or proceedings or in any other publication, or offered for publication elsewhere in substantially the same or abbreviated form, either in print or electronically.
Conflict of Interest
The authors declare that there is no conflict of interest.
Funding Sources
There was no direct funding for publication.
References
- Chennappa, G., Sreenivasan, M. Y., & Nagaraja, H. Azotobacter salinestris: a novel pesticide-degrading and prominent biocontrol PGPR bacteria. Microorganisms for Green Revolution: Microbes for Sustainable Agro-ecosystem, 2018;2:23-43.
CrossRef - Moneke AN, Okpala GN, Anyanwu CU. Biodegradation of glyphosate herbicide in vitro using bacterial isolates from four rice fields. Afr J Biotechnol. 2010; 9:4067-74.
- Schoffer, J.T., Solari, F., Petit-dit-Grézériat, L..The downside of copper pesticides: An earthworm’s perspective. Environ Sci Pollut Res 2024;31,16076–16084 https://doi.org/10.1007/s11356-024-32078-7
CrossRef - Sharma, S., Sharma, A., Bala, R., Sharma, I., & Sharma, A. Pesticide biology in soil: Sorption, leaching, and accumulation. In Pesticides in a Changing Environment 2024;49-66). Elsevier.
CrossRef - Sahoo, B., & Chaudhuri, S. Removal of lindane in liquid culture using soil bacteria and toxicity assessment in human skin fibroblast and HCT116 cell lines. Environmental Technology, 2023;44(9), 1213-1227.
CrossRef - Bhattacharyya PN, Jha DK. 2012 Plant Growth-Promoting Rhizobacteria (PGPR): emergence in agricultureWorld Microbiol Biotechnol.; 2021; 28(4): 1327-50.doi: 10.1007/s11274-011-0979-9
CrossRef - Romeh A. Remedial potential of Plant Growth Promoting Rhizobacteria (PGPR) for pesticide residues: recent trends and future challenges. Pestic Biorem: 2022;381-97.
CrossRef - Huang XD, El-Alawi Y, Penrose DM, Glick BR, Greenberg BM 2004. A multi-process phytoremediation system for removal of polycyclic aromatic hydrocarbons from contaminated soils. Environ Pollut.;130(3):465-76. doi: 1016/j.envpol.2003.09.031,
CrossRef - Jiang CY, Sheng XF, Qian M, Wang Isolation and characterization of a heavy metal-resistant Burkholderiasp. from heavy metal-contaminated paddy field soil and its potential in promoting plant growth and heavy metal accumulation in metal-polluted soil. Chemosphere. 2008;72(2):157-64. doi: 10.1016/j.chemosphere.2008.02.006.
CrossRef - Cycoń M, Mrozik A, Piotrowska-Seget Z. Bioaugmentation as a strategy for the remediation of pesticide-polluted soil: a review. Chemosphere .2017;172: 52-71.doi: 1016/j.chemosphere.2016.12.129
CrossRef - Pikovskaya’s RI. Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. 1948;17:362-70.
- Yaghoobi Khanghahi M, Pirdashti H, Rahimian H, Nemat Zadeh G, Ghajar Sepanlou M. Potassium Solubilising Bacteria (KSB) isolated from rice paddy soil: from isolation, identification to K use efficiency. Symbiosis 2018;76(1):13-23. doi: 1007/s13199-017-0533-0.
CrossRef - Dhaked BS, TriveniS, ReddyRS, PadmajaG. Isolation and screening of potassium and zinc solubilizing bacteria from different rhizosphere soil. Int J Curr Microbiol Appl Sci.; 2017;6(8):1271- doi: 10.20546/ijcmas.2017.608.154.
CrossRef - Mazumdar D, SahaSP, GhoshS. Isolation, screening and application of a potent PGPR for enhancing growth of Chickpea as affected by nitrogen level. 2020.;26(4):333-50. doi: 10.1080/19315260.2019.1632401.
CrossRef - Patel TS, Desai PB. Isolation and screening of PGPR from rhizospheric and non rhizospheric soil of Bt-cotton. Indo-2015;3: B1110-20.
- Singh RP, Jha P, Jha PN. The plant-growth-promoting bacterium Klebsiella SBP-8 confers induced systemic tolerance in wheat (Triticum aestivum) under salt stress. JPlantPhysiol.; 2015; 184:57-67. doi: 10.1016/j.jplph.2015.07.002
CrossRef - Bakthavatchalu S, Sivakumar, Sullies. Identification of multi-trait PGPR isolates and evaluation of their potential as biocontrol agents. Acta BiolInd.; 2012;1(1):61-7.
- Nayak, Tanmaya, Ananta N. Panda, Khushbu Kumari, Tapan Kumar Adhya, and Vishakha Raina. Comparative genomics of a paddy field bacterial isolate Ochrobactrum sp. CPD-03: analysis of chlorpyrifos degradation potential.” Indian Journal of Microbiology2020;60: 325-333.
CrossRef - Nwachukwu, B. C., Babalola, O. O., & Hassen, A. I. Rhizosphere competence and applications of plant growth-promoting rhizobacteria in food production-a review. Scientific African, 2024;23 e02081
CrossRef - Parmar P, Sindhu S. The novel and efficient method for isolating potassium solubilizing bacteria from rhizosphere soil. GeomicrobiolJ. 2019;36(2):130-
CrossRef - Mohite B.Isolation and characterization of Indole Acetic Acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. JSoilSciPlantNutr.; 2013; 13:638- doi: 10.4067/S0718-95162013005000051.
CrossRef - Ataikiru TL, OkerentugbaPO, OkpokwasiliGC;Identification of carbofuran and paraquat degrading microorganisms from South Asian. JResMicrobiol. 2020; 7:40-52.
CrossRef - Farhan M, Ahmad M, Kanwal A, ButtZA, KhanQF, RazaSA, Biodegradation of chlorpyrifos using isolates from contaminated agricultural soil, its kinetic studies. ; 2021; 11(1):10320. doi: 10.1038/s41598-021-88264-x
CrossRef - Mali H, Shah C, Patel DH, Trivedi U, Subramanian RB.Degradation insight of organophosphate pesticide chlorpyrifos through novel intermediate 2, 6-dihydroxypyridine by Arthrobacter sp. HM01. BioresourBioprocess.; 2022;9(1):1-14.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





