Manuscript accepted on : 20-03-2024
Published online on: 28-03-2024
Plagiarism Check: Yes
Reviewed by: Dr. Rakesh U. Shelke
Second Review by: Dr. Chetana SUVALKA
Final Approval by: Dr. Fernando José Cebola Lidon
Debleena Chatterjee1 , Ipsita Chanda2
, Ipsita Chanda2 and Archana Pan1*
and Archana Pan1*
1Department of Bioinformatics, Pondicherry University, Pondicherry, India.
2Department of Zoology, S.A. Jaipuria College, Kolkata, West Bengal, India.
Corresponding Author E-mail: archana@bicpu.edu.in
DOI : http://dx.doi.org/10.13005/bbra/3231
ABSTRACT: Enterobacter huaxiensis, a novel gram-negative bacterium of the family Enterobacteriaceae, was recovered from the blood of the patients at West China Hospital. The present study aims to predict potential lead molecules against the identified virulence-associated antibiotic-resistant protein for drug designing. Two virulence-associated antibiotic-resistance proteins belonging to the OqxAB efflux protein family of the RND superfamily were identified in the pathogen using bioinformatics tools/databases. Based on the structure prediction by homology modeling and validations, the RND transporter permease subunit, OqxB was selected as the potential target for lead identification. The binding pocket of the target protein was calculated using CASTp. A total of 204 phytochemicals were screened virtually to obtain compounds that had better binding affinity, drug-likeness and pharmacokinetic potential to be used as safe ligands against the target protein. Among these, Chrysoeriol, Isopimaric acid, Baicalein and Biochanin A were found to be within the permissible range of Lipinski rule of five for drug-likeness, possessing better ADMET properties, and lower target-protein binding energy (less than -8.0 kcal/mol). Ligand-protein docking showed stable non-covalent interactions between active site residues and ligands. Thus, these compounds may be considered potential inhibitors of the target protein that may inactivate the efflux pump and restore antibiotic sensitivity.
KEYWORDS: Antimicrobial resistance; E. huaxiensis; Efflux pump; Phytochemicals; Virulence
Download this article as:| Copy the following to cite this article: Chatterjee D, Chanda I, Pan A. Screening of Potential Phytochemicals for the Identified Candidate Virulence Proteins in Enterobacter huaxiensis. Biotech Res Asia 2024;21(1). |
| Copy the following to cite this URL: Chatterjee D, Chanda I, Pan A. Screening of Potential Phytochemicals for the Identified Candidate Virulence Proteins in Enterobacter huaxiensis. Biotech Res Asia 2024;21(1). Available from: https://bit.ly/43yERlX |
Introduction
Enterobacter huaxiensis is a novel gram-negative, motile, non-spore-forming and facultative anaerobe that belongs to the family Enterobacteriaceae. It was recovered from the blood of the patients at West China Hospital, Sichuan, PR China in 2017 and found resistant to multiple antibiotics.1 Enterobacter is generally present as a natural commensal microflora in the human gut and is usually not harmful. Few species of Enterobacter were reported to cause nosocomial infections in humans.2 Enterobacter infections in the bloodstream can cause serious consequences, such as bacteremia including sepsis, septic shock, and meningitis.3 The major causes of Enterobacter pathogenesis are associated with virulence factors as well as antimicrobial resistance. The virulence mechanism enables the microorganism to cause disease by evading the host’s immune response. Also, antimicrobial resistance hinders antimicrobial therapy facilitating the survival of microbial pathogens under adverse conditions.4 Moreover, some antimicrobial resistance factors are involved in causing direct virulence in the host.5 There are reports of drug-resistance and virulence roles of potential drug transporters of RND (AcrAB, AcrD and AcrEF system), MFS, MATE and ABC superfamilies in Enterobacter cloacae, Klebsiella pneumoniae and Salmonella enterica serovar Typhimurium and other gram-negative bacteria.6-9 The virulence factors and antimicrobial resistance are propagated using horizontal transfer of genes among different Enterobacter strains.5
The efflux pump system is a key mechanism of antibiotic resistance in gram-negative bacteria.10 This system expels antimicrobial agents out of the cell. Amongst the six families of bacterial efflux pumps, the most clinically significant efflux system in gram-negative bacteria belongs to the resistance nodulation division (RND) superfamily.11 Efflux system of the RND superfamily is composed of tripartite protein complexes comprising an inner membrane protein (IMP), an outer membrane protein (OMP) and a periplasmic adaptor protein (PAP).12 AcrAB-TolC in E. coli and MexAB-OprM in P. aeruginosa were extensively studied multidrug resistance RND type efflux protein in gram-negative bacteria.13 One of the ways of efflux pump inhibition is blocking the pump to avoid substrate binding at the active site.14
Natural plants are traditionally used worldwide for the treatment of various diseases owing to their rich traditional medicinal value. People in rural areas mostly rely on natural plants for common ailments, such as cough and cold, skin disease, gastrointestinal disorders, urinary problems, hepatic disease, and many others.15,16 About 25% of the most prescribed drugs are obtained from plants.17 Thus, plants are important resources for the development of new drugs. Phytochemicals, extracted from different parts of plants, such as roots, stems, leaves, and flowers are filtered and modified into a form that can successfully be used as antimicrobial agents.18 There is evidence that phytochemicals such as terpenoids, phenylpropanoids, flavonoids, coumarins, sterols, alkaloids, tannins, saponins, and polyphenols are biologically active and act as good antibacterial agents to combat multiple antibiotic-resistant bacterial infections.19,20 Moreover, phytochemicals are effective anti-oxidants and pose fewer side effects than modern-day chemical medication.21
Computational approaches to screening lead compounds facilitate the drug discovery process in a time- and cost-effective manner, bypassing the hazards of increasingly challenging analytical methods.22 Screening of phytochemical compounds has progressively become an important tool for modern drug discovery programs, particularly to combat the expanding antibiotic-resistant bacteria.23 The in silico methods encompass approaches, such as homology or another molecular modelling, virtual screening, candidate building, docking, pharmacokinetic, ADMET and drug likeliness prediction.24
Herein, this study is aimed to (i) identify the candidate virulence and antibiotic resistance proteins in Enterobacter huaxiensis, and, (ii) screen out phytochemicals as the lead compound against the selected target protein of E. huaxiensis for future development of the drug. The phytochemicals are mostly obtained from plants of tropical and subtropical distribution and are reported to have various medicinal uses.
Materials and Methods
Prediction of Virulence Associated proteins
The protein-coding genes and their corresponding protein sequences of the bacterium Enterobacter huaxiensis, were retrieved from the NCBI Genome database (https://www.ncbi.nlm.nih.gov/genome/ 74721?genome_assembly_id=431299). The hypothetical proteins were discarded from the protein sequence list. From this obtained protein sequence list, virulence factors (VF) that are likely to be associated with pathogenesis in this organism were identified using the MP3 tool and virulence factor database (VFDB).25,26 The proteins that showed virulence by both VFDB and MP3 were further considered for analysis. MP3 (http://metagenomics.iiserb.ac.in/mp3/index.php) is a tool that uses SVM and HMM principles to predict the virulence of a query sequence. VFDB (http://www.mgc.ac.cn/VFs/) is a pathogenomic platform that provides details on virulence factors present in an organism.
Additionally, RGI (Resistance Gene Identifier) in CARD (The Comprehensive Antibiotic Resistance Database) was used to determine whether the identified virulence factors were antibiotic-resistant.27 The CARD (https://card.mcmaster.ca/) is a resource of resistance genes and their associated antibiotics. RGI is a tool in CARD for the prediction of antibiotic resistance genes.
Structure Prediction and Validation
Homology modeling was performed for selected virulence factors using MODELLER (version 10.1, release date 12/03/2021, from https://salilab.org/modeller/).28 The templates for selected proteins were collected from RCSB PDB (https://www.rcsb.org/), an online repository for crystallographic structures of protein macromolecules. The modeled structures were validated with Ramachandran Plot, PROCHECK, ERRAT, PROVE and other tools from SAVES server (https://saves.mbi.ucla.edu/). Loop refinement was carried out using an online tool, MODLOOP (https://modbase.compbio.ucsf.edu/modloop/).
Active Site Prediction
CASTp (Computed Atlas of Surface Topography of Proteins) (http://sts.bioe.uic.edu/castp/index.html?3igg) is an online platform, which was used to predict the binding pocket cavities and active site residues, present in the modelled proteins.29
Collection of Phytochemicals
Phytochemicals, obtained from different parts of medicinal plants of tropical and subtropical origin were collected through a literature survey. Phytochemicals that have different medicinal uses including the inhibitory effects on the growth of bacteria were listed for the study.30-42
Molecular Docking Study
Protein preparation
Using PyMOL 2.4.0, the water molecules and other unwanted atoms/ligand molecules attached to the protein of interest were manually removed.43 Subsequently, the protein was processed using AutoDockVina Tools 1.5.6 (http://autodock.scripps.edu/resources/adt/) and converted into “.pdbqt” file format, which could be used for molecular docking study.44
Ligand preparation
The structures of all the selected phytochemicals were retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/) in “.sdf” format. Using PyMOL 2.4.0, these compounds were cleaned and converted into “.pdb” format. The compounds obtained from PubChem having 2D structures were cleaned using MarvinSketch 19.17 (http://www.chemaxon.com/) and converted into a 3D structure format. Those were then converted into “.pdb” file format using PyMOL 2.4.0. Finally, by using AutoDockVina Tools 1.5.6, compounds were processed as per necessary protocols and saved in “.pdbqt” file format for further use in molecular docking studies.
Grid Box generation
Within AutoDockVina, the active site residues obtained from CASTp were selected to highlight portions of the protein where the ligand is to be bound. Accordingly, a grid box is generated (with a default spacing of 0.375 Å) in such a way that it covers the entire portion of all the binding cavities within the protein, and the coordinates are noted.
Docking and Binding Energy analysis
AutoDockVina 1.5.6 (https://vina.scripps.edu/) is based on the iterated local search global optimizer algorithm, which uses the Broyden-Fletcher-Goldfarb-Shanno (BFGS) method, an efficient quasi-Newton method for local optimization. The molecular docking study was performed with an exhaustiveness factor set to 100, to minimize any possibility of residues being left out. After each prepared phytochemical was docked with the prepared protein, the respective conformation file was generated in “.pdbqt” file format and was considered for further analysis. The log file, corresponding to the same containing the molecular docking score was generated in “.txt” file format.
Prediction of ADMET properties
Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties were predicted for the selected phytochemicals having the best docking scores. This was done using the pkCSM webserver (http://biosig.unimelb.edu.au/pkcsm/) as well as the Swiss-ADME webserver (http://www.swissadme.ch/) for better interpretation of results.
Protein-ligand interaction analysis
The conformation files of top-scoring phytochemicals, obtained from the molecular docking study were opened using PyMOL and were made to complex with the respective target protein. The complex of ligand and protein was saved in “.pdb” file format. The protein-ligand interactions of the complexes were then analyzed, using the Protein-Ligand Interaction Profiler (PLIP) web server (https://plip-tool.biotec.tu-dresden.de/plip-web/plip/index). PLIP uses a four-step algorithm for the detection and visualization of relevant interactions between ligands and receptors and provides a text file containing all details regarding various types of interaction and the exact sites of their occurrence.45
Results
Identification of virulence- associated target proteins
The Enterobacter huaxiensis genome comprises 4401 annotated protein-coding genes (data collated in July 2021). The annotated dataset was explored for putative virulence factors using the MP3 tool and VFDB. A total of 1166 proteins were predicted as virulence factors using the MP3 tool and about 113 proteins showed significant hits to the virulence factor database (VFDB). The datasets obtained using the MP3 tool and VFDB were then compared. About 43 virulent proteins were found in both datasets and were listed for further study. Moreover, the primary dataset of 4401 annotated proteins was searched for antimicrobial resistance using RGI from CARD. A total of 21 proteins exhibited antimicrobial resistance properties and were listed for the study. These datasets of virulence and antimicrobial resistance proteins were compared to screen out the proteins that have roles both in virulence and antimicrobial resistance. Amongst these, two proteins were found in both datasets indicating virulence and antimicrobial resistance activities (Fig 1). Table 1 shows that these proteins are efflux pump proteins that belong to the RND superfamily and exercise resistance via the antibiotic efflux mechanism. Homology searching was carried out to confirm that these proteins had no human homologs, and were selected for structure prediction.
 |
Figure 1: Selection of virulence-associated antibiotic-resistance proteins |
Table 1: List of virulence-associated antibiotic-resistant proteins and functions.
| ORF_ID | Length | Function | Drug Class | Resistance Mechanism | AMR Gene Family |
| WP_119934747.1 | 1051 | Multidrug efflux RND transporter permease subunit, OqxB | Fluoroquinolone; Tetracycline antibiotics. | Antibiotic efflux | Resistance-nodulation cell division (RND) antibiotic efflux pump |
| WP_119934748.1 | 391 | Efflux RND transporter periplasmic adaptor subunit, OqxA | Fluoroquinolone; Glycylcycline; Tetracycline; Diaminopyrimidine; Nitrofuran. | Antibiotic efflux | Resistance-nodulation cell division (RND) antibiotic efflux pump |
Structure prediction and validation
Homology models of the selected efflux proteins of E. huaxiensis were constructed ….. Protein Data Bank (PDB). Two templates were selected for the two proteins based on the sequence identity and significant e-value (Table 2). Sequence identity above 30% is a relatively good predictor of the expected accuracy of a model.46 It was found that a multi-drug resistant efflux pump of Campylobacter jejuni (5LQ3) showed 40.62% sequence identity and e-value 0.0 with efflux RND transporter permease subunit, OqxB (WP_119934747.1). Similarly, an RND-membrane transport multi-drug efflux pump of Escherichia coli (5V5S) showed 33.51% sequence identity and e-value 1e-49 with efflux RND transporter periplasmic adaptor subunit, OqxA (WP_119934748.1). In the present study, the template (5LQ3) with higher sequence identity and e-value score was selected as the target protein for further analysis. This template was used to build a homology model of efflux RND transporter permease subunit, OqxB (WP_119934747.1). The 3D structure of the target efflux protein was constructed using homology modeling and Loop refinement was carried out by MODLOOP (Fig 2).
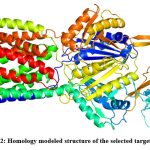 |
Figure 2: Homology modeled structure of the selected target protein.Click here to view Figure |
Table 2: Template selection for Homology Modeling of the protein
| Virulence Factor (Accession number) | Length | PDB ID of template, chain | Query cover | Percent Identity | E-value | Protein details | Organism name |
| WP_119934747.1 | 1051 | 5LQ3,A chain | 98% | 40.62% | 0.0 | Structure and transport dynamics of the multidrug efflux pump. | Campylobacter jejuni |
| WP_119934748.1 | 391 | 5V5S,D chain | 97% | 33.51% | 1e-49 | Multi-drug efflux; Membrane transport; RND superfamily; Drug resistance | Escherichia coli |
The structure was validated using Ramachandran Plot that evaluates dihedral angles- phi (ɸ) and psi (Ψ) of amino acid residues to predict the allowed and disallowed conformations of the target protein structure. The plot indicated 92.3% residues in the most favored regions, 6.5% residues in the allowed region and none of the residues in the disallowed region. The homology model was further verified using ERRAT, PROCHECK, PROVE, and other tools from SAVES server. ERRAT scored the overall quality factor for non-bonded atomic interaction, giving a score of 76.385 which represents the credibility and high quality of the predicted model. PROCHECK verified the deviations of main chain parameters like peptide bond planarity, non-bonded interactions, Cα tetrahedral distortion, main chain hydrogen bond energy, and overall G factor from the template lie within the favorable range. PROVE verified the deviations of the atomic volumes (Z-score RMS) from the standard that decreases as the resolution improves reflecting the accuracy of the model. Figure 3 shows the structure validation by Ramachandran plot, PROCHECK and ERRAT programs. Further, the superimposition of the target protein with the template predicted the RMSD value of 0.169 Å indicating that the modeled target structure was reliable.
 |
Figure 3: Structure validation of the target modeled protein. (A) Ramachandran plot showing 92.8% residues in the allowed region; (B) PROCHECK validation plot for main chain parameters.Click here to view Figure |
Identification of binding pocket of virulence factor
CASTp 3.0 was used for an analysis of the ligand binding pocket of the predicted target protein. A 1.4 Å radius probe of the internal cavity surface volume of the ligand binding pocket was calculated. The binding pocket was large with a surface area of 4052.809 sq. Å and volume 4662.109 cubic Å. PyMOL was used to analyze the results obtained through CASTp so the active sites could be better understood. Upon thorough examination, it was found that the binding pocket contains several hydrophobic, aromatic and hydrophilic residues (P52, V83, V91, P136, L624, F669, I671, I832, G833, G684, F626, P630, L758, G589, G759, Y79, Y688, Y278, T93, K585, T632, S683, D834, E588, N628, Q755, Q778, S86, D87, N631, S760, Q778, D818, D867) to which ligands can potentially bind.
Virtual screening of phytochemicals
A dataset of 204 phytochemicals obtained from 42 plant species were listed for the present study (Table S1). These phytochemicals belong to various classes, such as indoles, steroids, organic acids, phenols, benzoates, flavonoids, alkaloids, alkanes, terpenoids etc. compounds. The listed phytochemicals were subjected to a virtual screening protocol to select the druggable compounds. Identification of target protein structure and binding pocket helps in the identification of potent lead molecules through ligand-protein interaction.47 The phytochemicals were docked against the target protein as the first phase of the selection filter. The ligands with higher binding affinity for the target protein were considered for further analysis. About 73 phytochemicals with binding energy less than -8 kcal/mol were selected for the next phase of screening. The threshold was selected based on the previously reported RND efflux pump inhibitor.48 These compounds were tested for Lipinski’s rule of five (Ro5) for drug-likeness. According to Ro5, a compound is an orally active drug if its physicochemical properties lie within the established range with no more than one violation (molecular weight ≤ 500 Da, hydrogen bond donors ≤ 5, hydrogen bond acceptors ≤ 10, and an octanol-water partition coefficient LogP ≤ 5.).49 The compounds were also tested for Veber’s rule and Ghose filter violations to select the compounds with druggable properties. Veber’s rule states that a good oral bioavailable compound should have number of rotatable bonds ≤10 and topological polar surface area ≤140 sq Å.50 Ghose filter defines that a drug-like compound possesses parameters, such as LogP in between 5.6 and -0.4, molecular weight in between 480 and 160, molar refractivity in between 130 and 40, and total number of atoms in between 70 and 20.51 Out of 73 compounds, 28 were selected based on the screening through Ro5, Veber’s rule and Ghose filter. These phytochemicals were further subjected to ADMET analysis. ADMET-based drug screening is the crucial and most important step in drug discovery to identify the compounds that have biochemical properties safe for a clinical trial. The compounds with no toxigenicity, mutagenicity, and carcinogenicity are considered safe drug candidates. Moreover, these compounds were also tested for their water solubility, gastrointestinal (GI) absorption, P-glycoprotein substrate, Blood-Brain Barrier (BBB) permeability, and cytochrome P450 metabolism. The ADMET analysis finally screened out four compounds, selected as the lead (L1-L4) for the target efflux protein of E. huaxiensis (Table 3). Figure 4 shows the virtual screening flow of the study.
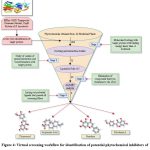 |
Figure 4: Virtual screening workflow for identification of potential phytochemical inhibitors of the Efflux RND transporter permease subunit, OqxB protein of E. huaxiensis |
 |
Table 3: List of selected phytochemicals, sources, and chemical descriptions. |
Drug-likeness and pharmacokinetic parameters
Table 4 shows the physicochemical parameters of drug-likeness properties of the selected compounds. According to the result, compounds L1, L3, and L4 showed no violation of Lipinski’s rule of five, Veber’s rule, and Ghose filter. The compound, L2 showed one violation of the Lipinski rule filter. The pharmacokinetic properties of the selected compounds were analyzed by ADMET parameters (Table 5). According to the result, the water solubility of the compounds ranges from moderately soluble to soluble according to the three solubility classes. GI absorption was high for all compounds and the LogKp value for skin permeability varied between -4.38 to -5.93. L3 was the only compound that was BBB permeable. All four compounds were non-substrate of P-glycoproteins (Pgp). All compounds, except L2, were inhibitors of cytochrome P450 enzyme isoforms, CYT1A2, CYT2D6 and CYT3A4. L2 inhibited CYT2C19 and CYT2C9 isoforms only. The total clearance (CLtot) value of the compounds fell between the range of 0.247 to 0.717. The range of LD50 toxicity was between 1.851 to 2.337. The compounds were predicted to be non-mutagenic and non-hepatotoxic.
Table 4: Physicochemical properties of selected phytochemicals.
|
Phytochemical |
Mol wt (g/mol) | LogP | HBA | HBD | RB | TPSA (Å2) | Lipinski rule of violation | Veber’s rule of violation |
Ghose filter violation |
| Chrysoeriol | 300.26 | 2.58 | 6 | 3 | 2 | 100.13 | 0 | 0 | 0 |
| Isopimaric acid | 302.45 | 5.20 | 1 | 1 | 2 | 37.3 | 1 | 0 | 0 |
| Baicalein | 270.24 | 2.57 | 5 | 3 | 1 | 90.90 | 0 | 0 | 0 |
| Biochanin A | 284.26 | 2.87 | 5 | 2 | 2 | 79.90 | 0 | 0 | 0 |
HBA= H-bond acceptor, HBD= H-bond donor, RB= Rotatable bond, TPSA=Topological Polar Surface Area
Table 5: Pharmacokinetic properties of selected phytochemicals.
| ADMET | Parameters | Chrysoeriol | Isopimaric acid | Baicalein | Biochanin A |
| Water Solubility | ESOL Class | Moderately Soluble | Moderately Soluble | Moderately Soluble | Soluble |
| Ali Class | Moderately Soluble | Moderately Soluble | Moderately Soluble | Moderately Soluble | |
| Silicos-IT class | Moderately Soluble | Soluble | Moderately Soluble | Moderately Soluble | |
| Absorption | GI absorption | High | High | High | High |
| Skin permeability LogKp (cm/s) | -5.93 | -4.38 | -5.7 | -5.91 | |
| Distribution | BBB permeability | No | Yes | No | No |
| Pgp substrate | No | No | No | No | |
| Metabolism | CYP1A2 inhibitor | Yes | No | Yes | Yes |
| CYP2C19 inhibitor | Yes | Yes | No | No | |
| CYP2C9 inhibitor | No | Yes | No | No | |
| CYP2D6 inhibitor | Yes | No | Yes | Yes | |
| CYP3A4 inhibition | Yes | No | Yes | Yes | |
| Excretion | CLtot (log ml/min/kg) | 0.597 | 0.717 | 0.252 | 0.247 |
| Toxicity | LD50 (mol/kg) | 2.337 | 1.881 | 2.325 | 1.851 |
| Mutagenicity | No | No | No | No | |
| Hepatotoxicity | No | No | No | No |
Interaction of E. huaxiensis target protein with selected ligands
Table 6 displays the 2D chemical structures of the top 4 phytochemical ligands (L1-L4) of E. huaxiensis efflux protein. Based on PubChem classification, L1, L3, and L4 are flavonoids and L2 is a terpenoid.
Phytochemical L1 (Chrysoeriol) is a 4,5,7-trihydroxy-3′-methoxyflavone, produced by the herb, Plectranthus glandulosus. In plants, flavonoids act as defense chemicals.52 It also possesses several medicinal uses in humans.53 In the study, the docking score of Chrysoeriol with the specific target was -8.8 kcal/mol. In the ligand-protein complex, three hydroxyl groups of the compound showed interactions with Q46, K81, and Y688 residues by the hydrogen bonds. Ligand binding was further stabilized by the hydrophobic interactions with active site residues (Fig 5, Table 6).
Phytochemical L2 (Isopimaric acid) is a diterpenoid (a carbotricyclic compound and a monocarboxylic acid), produced by the plant, Pinus brutia var. eldarica. It acts as an ionophore in plants by increasing the permeability of specific ions through biological membranes. In humans, it exhibits diverse pharmacological activities.54 The ligand-protein binding energy of Isopimaric acid in this study was -8.8 kcal/mol. H-bond interaction was absent. Ligand binding was dominated by hydrophobic interactions with the binding site residues (Fig 5, Table 6).
Phytochemical L3 (Baicalein) is a 5,6,7-trihydroxyflavon, a plant metabolite that has several medicinal uses in prostate cancer, Alzheimer’s disease, and dementias, and acts as an antioxidant, anti-inflammatory, antineoplastic, antibacterial, and antifungal agents.55 It was first isolated from the leaves of Thymus vulgaris (garden thyme) and roots of Scutellaria baicalensis.56 The target protein binding energy of Baicalein in the study was -8.3 kcal/mol. In the protein-ligand complex, the two hydroxyl groups of the compound interact with the residues Y79, Y688, and D867. Ligand binding further engaged several hydrophobic interactions with binding pocket residues (Fig 5, Table 6).
Phytochemical L4 (Biochanin A) is a 5,7-dihydroxy-4-methoxyisoflavone found in the plant, Lupinus argenteus. It is a phytoestrogen and has putative benefits in dietary cancer prophylaxis. The ligand-protein binding energy was -8.3 kcal/mol. The hydroxy groups generated H-bonds with T632 and G833 of the active site residues. The ligand-protein complex was engaged in several hydrophobic bonds with active site residues (Fig 5 and Table 6).
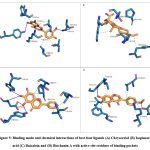 |
Figure 5: Binding mode and chemical interactions of best four ligands (A) Chrysoeriol (B) Isopimaric. |
Table 6: Binding energy and interactions between selected phytochemicals and target protein binding pocket.
| Ligand | Binding Energy (kcal/mol) | RMSD | Interacting Residues | |
| Hydrophobic Interactions | Hydrogen Bonds | |||
| Chrysoeriol | -8.8 | 0.0 | Y79, V83, V91, T93 | Q46, K81, Y688 |
| Isopimaric acid | -8.8 | 0.0 | K81, V91, T93, P136, F626 | – |
| Baicalein | -8.3 | 0.0 | V83, V91, T93, P136 | Y79, Y688, D867 |
| Biochanin A | -8.3 | 0.0 | K585, L624, F669, I671, I832, | T632, G833 |
Discussion
Enterobacter huaxiensis is a novel gram-negative bacterium, recovered from the blood of the infected patients at West China Hospital. In the last few decades, this genus has gained clinical importance as a nosocomial pathogen.1 E. huaxiensis exhibits resistance to multiple antibiotics.1 A total of 4401 annotated protein-coding genes of E. huaxiensis were searched for virulence-associated and antibiotic-resistance proteins using the VFBD database, MP3 tool, and CARD database, respectively. Two proteins were identified as possessing both properties of conferring virulence as well as exhibiting antibiotic resistance. The proteins belonged to the OqxAB efflux pump of the RND superfamily, one with multidrug efflux RND transporter permease subunit, OqxB (WP_119934747.1), and the other with efflux RND transporter periplasmic adaptor subunit, OqxA (WP_119934748.1). Homology-modeled structures of the selected proteins were prepared and structures were validated. Templates with sequence identity above 30% were used for the expected accuracy of the model.46 However, based on the higher percentage identity of the template, the multidrug efflux RND transporter permease subunit, OqxB (WP_119934747.1) was finally selected as the target protein for further study. OqxB is an inner membrane protein of the OqxAB efflux pump of the RND superfamily and is homologous to efflux pumps of the RND family in other bacterial species (AcrB, MexB, CmeB AdeB, and MtrD).57 OqxAB is a chromosome and/or plasmid-encoded multidrug efflux pump that is flanked by a transposable element (IS26), which facilitates the dissemination of antimicrobial resistance via horizontal transfer.57 Ramachandran Plot showed that >90% of residues of the modeled structure fell in the favored region, which is considered to be the measure of a good quality model (Fig 1).58 The prediction of binding pockets of the modeled structure was calculated using CASTp. The binding pocket is large with several active site residues predominated by Pro, Gly, Leu, Val, Phe, Tyr, Thr, Ser, Asp, and Gln.
A total of 204 phytochemicals belonging to various chemical classes were obtained from different plants of tropical and subtropical distribution (Table S1). The phytochemicals were screened through a virtual screening protocol to obtain probable ligands for the target protein of E. huaxiensis (Fig 3). Virtual screening method of phytochemicals predicted four ligands that include Chrysoeriol, Isopimaric acid, Baicalein, and Biochanin A. Among these, Chrysoeriol, Baicalein, and Biochanin A belong to the class flavonoid and Isopimaric acid is a terpenoid. The docking score or binding energy is a function of the binding affinity of the ligand to the target protein. The lower binding energy indicates a higher affinity for ligand-protein binding. In the present study, the ligands exhibit binding energy lower than the threshold (less than -8.0 kcal/mol) (Table 5). This indicates higher binding affinity of the ligands with the binding pocket active site residues of the target. The interactions of the ligands with active site residues significantly affect the activity of the target protein and result in inhibition of their activity.
A compound may be considered a potential candidate drug if it possesses drug-like properties and optimal pharmacokinetic characteristics. In the present study, the ligands were tested for drug-likeness and pharmacokinetic properties, by using pkCSM ADMET and Swiss-ADME web servers. All the compounds were considered drug-like, having parameters within the range established by Lipinski’s rule of five (Ro5), Veber’s rule, and Ghose filter along with violations (Table 4).49-51 The greater LogP value of Isopimaric acid indicates its lipophilic property implying that the compound can diffuse through the cell membrane to be bioavailable. Solubility is that physiochemical property that determines the absorption, distribution, and formulation of the drug.59 In this study, all compounds were water-soluble. GI absorption is an important criterion for an oral drug because the primary site of absorption of orally administered drugs is the GI tract.60 The four compounds in the present study were found to be well-absorbed by the GI tract. Skin permeability is the predictor of transdermal delivery of drugs. It depends on the linear correlation with molecular size and lipophilicity. The more negative the LogKp value, the less skin permeant the molecule.60 In the present study, Chrysoeriol, Baicalein, and Biochanin A had higher negative LogKp values and were less skin permeant than Isopimaric acid. BBB permeability indicates the property of a drug to cross the Blood-Brain Barrier and produce side effects. However, it also improves the efficacy of the drugs which show their pharmacological activities within the brain.59 In the present study, only Isopimaric acid had BBB permeability. This might be correlated to the lipophilic property of the drug.61 P-glycoprotein (Pgp) is an important ATP-binding cassette (ABC) transporter prominent in the GI wall and brain that extrudes toxins and xenobiotics out of the cell and determines the distribution of the compounds within the body.60 In the present study, all ligands were non-substrates of Pgp. Cytochrome P450 is an important enzyme, present mainly in the liver. It helps in the detoxification and oxidation of drugs to facilitate their excretion. However, many drugs are deactivated by cytochrome P450 enzyme, while some drugs are also activated by them. Among others, two main isoforms, CYP2D6 and CYT3A4 are responsible for drug metabolism.60 The inhibitors of Cytochrome P450 enzyme thus play a dramatic role in altering the pharmacokinetic properties of those drugs. In this study, all compounds except Isopimaric acid are inhibitors of CYP2D6 and CYT3A4. These compounds inhibit the enzymes that metabolize the drugs causing inactivation, in addition to the potential to increase drug levels in the blood and cause adverse drug reactions.61 Chrysoeriol inhibits most of the cytochrome P450 enzyme isoforms. Regarding overall clearance of the drug, Isopimaric acid and Chrysoeriol have better total clearance rates indicating a lower risk of accumulation and toxicity.59 Based on the LD50 value, Isopimaric acid, and Biochanin A have toxicities higher than other compounds and their safety requires evaluation.61 The compounds under study have no mutagenic/ carcinogenic potential based on the Ames test observation and do not associate with drug-induced liver injury which is considered as the major safety concern for drug development.62
Conclusion
In summary, two multidrug efflux RND transporter proteins, one with permease OqxB and the other with adaptor subunit, OqxA were identified as virulence-associated antibiotic-resistance proteins in Enterobacter huaxiensis, a novel bacterium of family Enterobacteriaceae that was recovered from the blood of the patients at West China Hospital in 2017. Based on structure prediction and validation of the identified proteins, multidrug efflux RND transporter permease subunit, OqxB was selected as the potential target for lead development. A total of 204 phytochemicals acquired from different plant sources were virtually screened to obtain putative ligands. The binding affinity, physicochemical, and pharmacokinetic filters were applied to all phytochemicals to select druggable compounds. Four phytochemicals (Chrysoeriol, Isopimaric acid, Baicalein, and Biochanin A) demonstrated drug-like properties that were allowed by Lipinski’s rule of five, Veber’s rule and Ghose filter, and exhibited better ADMET characteristics. These compounds have higher ligand-protein binding affinity, exhibiting stable non-covalent interactions with the active site residues in binding pockets of target protein. Hence, these four compounds can be considered as the potential inhibitors of efflux RND transporter permease subunit, OqxB that may inactivate RND efflux and rejuvenate the sensitivity of the antibiotics.14 However, in vitro and in vivo experimental validations are required before consideration of these compounds as potential drugs for human administration.
Acknowledgement
The authors acknowledge the infrastructural support of the Department of Bioinformatics, Pondicherry University, Pondicherry, India. DC is thankful to BP and PG for technical assistance at the beginning of the work.
Conflict of Interest
No conflict of interest.
Funding statement
There is no funding sources
Author’s Contribution
Debleena Chatterjee: Data collection, analysis and interpretation of results, draft manuscript preparation.
Ipsita Chanda: Formal analysis, reviewed the results, edited and finalised the manuscript.
Archana Pan: Study conception,formal analysis, reviewed the results, edited and finalised the manuscript.
All authors have read and approved the final version of the manuscript.
Data Availability Statement
Not Applicable
Ethics Approval Statement
Not Applicable
References
- Wu W, Wei L, Feng Y, Kang M and Zong Z. Enterobacter huaxiensis sp. nov. and Enterobacter chuandaensis sp. nov., recovered from human blood. Int. J. Syst. Evol. Microbiol., 2019 Mar; 69(3): 708-714. doi: 10.1099/ijsem.0.003207. PMID: 30614784. Ramirez D and Giron M. Enterobacter Infections. StatPearls Publishing LLC.: Treasure Island; 2023.
CrossRef - Kang C. I, Kim S. H, Park W. B, Lee K. D, Kim H. B, Oh M, Kim E. C and Choe K. W. Bloodstream Infections Caused by Enterobacter Species: Predictors of 30-Day Mortality Rate and Impact of Broad-Spectrum Cephalosporin Resistance on Outcome. Clinical Infectious Diseases, 39(6), 812–818. https://doi.org/10.1086/423382
CrossRef - Bujňáková D, Puvača N and Ćirković I. Virulence Factors and Antibiotic Resistance of Enterobacterales. Microorganisms 2022; 10(8):1588; doi: 10.3390/microorganisms10081588.
CrossRef - Alverez-Ortega C, Olivares J and Martinez J. L. RND multidrug efflux pumps: what are they good for? Front. Microbiol. 2013;4; doi: https://doi.org/10.3389/fmicb.2013.00007.
CrossRef - Kondo K, Kawano M and Sugai M. Distribution of Antimicrobial Resistance and Virulence Genes within the Prophage-Associated Regions in Nosocomial Pathogens. mSphere 2021;6(4); doi: https://doi.org/10.1128/msphere.00452-21.
CrossRef - Pérez A, Poza M, Fernández A, del Carmen Fernández M, Mallo S, Merino M, Rumbo-Feal S, Cabral M. P and Bou G. Involvement of the AcrAB-TolC Efflux Pump in the Resistance, Fitness, and Virulence of Enterobacter cloacae. Antimicrobial Agents and Chemotherapy 2012; 56(4):2084-2090; doi:https://doi.org/10.1128/aac.05509-11
CrossRef - Fernando D. M, Kumar A. Resistance-Nodulation-Division Multidrug Efflux Pumps in Gram-Negative Bacteria: Role in Virulence. Antibiotics 2013; 2(1):163-181; doi:https://doi.org/10.3390/antibiotics2010163
CrossRef - Nishino K, Latifi T, Groisman E. A. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Molecular Microbiology 2006; 59(1):126-141; doi:https://doi.org/10.1111/j.1365-2958.2005.04940.x
CrossRef - Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea J. A, Albertí S. Klebsiella pneumoniae AcrAB Efflux Pump Contributes to Antimicrobial Resistance and Virulence. Antimicrobial Agents and Chemotherapy 2010; 54(1):177-183; doi:https://doi.org/10.1128/AAC.00715-09
CrossRef - Soto S. M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013; 4(3):223–229; doi: 10.4161/viru.23724
CrossRef - Fernando D. M and Kumar A. Resistance-Nodulation-Division Multidrug Efflux Pumps in Gram-Negative Bacteria: Role in Virulence. Antibiotics (Basel) 2013; 2(1):163–181; doi: 10.3390/antibiotics2010163
CrossRef - Alav I, Kobylka J, Kuth M. S, Pos K. M, Picard M, Blair J. M. A and Bavro V. N. Structure, Assembly, and Function of Tripartite Efflux and Type 1 Secretion Systems in Gram-Negative Bacteria. Chem. Rev. 2021; 121(9):5479–5596; doi: 10.1021/acs.chemrev.1c00055.
CrossRef - Ma M, Lustig M, Salem M, Mengin-Lecreulx D, Phan G and Broutin I. MexAB-OprM Efflux Pump Interaction with the Peptidoglycan of Escherichia coliand Pseudomonas aeruginosa. Int J Mol Sci. 2021; 22(10):5328; doi: 10.3390/ijms22105328.
CrossRef - Sharma A, Gupta V. K and Pathania R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J Med Res 2019; 149(2):129-145; doi: 10.4103/ijmr.IJMR_2079_17.
CrossRef
- Rahman M. H, Roy B, Chowdhury G. M, Hasan A and Saimun M. S. R. Medicinal plant sources and traditional healthcare practices of forest-dependent communities in and around Chunati Wildlife Sanctuary in southeastern Bangladesh. Environmental Sustainability 2022; 5(2):207–241; doi: https://doi.org/10.1007/s42398-022-00230-z.
CrossRef - Aqil F and Ahmad I. Antibacterial properties of traditionally used Indian medicinal plants. Methods and Findings in Experimental and Clinical Pharmacology 2007; 29(2):79; doi: 10.1358/mf.2007.29.2.1075347.
CrossRef - Houghton P. J. Old Yet New—Pharmaceuticals from Plants. Journal of Chemical Education 2001; 78(2); doi: 10.1021/ed078p175.
CrossRef - Banu K. S and Cathrine L. Phytochemicals, extracted from different parts of natural plants, such as roots, stems, leaves, and flowers are filtered and modified into a form that can successfully be used as antimicrobial agents. International Journal of Advanced Research in Chemical Science (IJARCS) 2015; 2(4):25-32.
- Gonelimali F. D, Lin J, Miao W, Xuan J, Charles F, Chen M and Hatab S. R. Antimicrobial properties and mechanism of action of some plant extracts against food pathogens and spoilage microorganisms. Front. Microbiol. 2018; 9; doi: https://doi.org/10.3389/fmicb.2018.01639
CrossRef - Nascimento G. G. F, Locatelli J, Freitas P. C and Silva G. L. Antibacterial activity of plant extracts and phytochemicals on antibiotic-resistant bacteria. Braz. J. Microbiol. 2000; 31(4); doi: 10.1590/S1517-83822000000400003.
CrossRef - Rajkumar S. R. J, Nadar M. S. A. M and Selvakumar P. M. Phytochemicals as a potential source for antimicrobial, anti-oxidant and wound healing – a review. Bioorganic & Organic Chemistry 2018; 2(2); doi: 10.15406/mojboc.2018.02.00058.
CrossRef - Hung C. L and Chen C. C. Computational Approaches for Drug Discovery. Drug Development Research 2014; 75(6):412–418; doi: 10.1002/ddr.21222.
CrossRef - Lage O. M, Ramos M. C, Calisto R, Almeida E, Vasconcelos V and Vicente F. Current Screening Methodologies in Drug Discovery for Selected Human Diseases. Mar Drugs. 2018; 16(8): 279; doi: 10.3390/md16080279.
CrossRef - Ekins S, Mestres J and Testa B. In silicopharmacology for drug discovery: methods for virtual ligand screening and profiling. Br J Pharmacol. 2007; 152(1): 9–20; doi: 10.1038/sj.bjp.0707305.
CrossRef - Liu B, Zheng D, Jin Q, Chen L and Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Research 2018; 47(D1):D687–D692; doi: 10.1093/nar/gky1080.
CrossRef - Gupta A, Kapil R, Dhakan D. B and Sharma V. K. MP3: A Software Tool for the Prediction of Pathogenic Proteins in Genomic and Metagenomic Data. PLoS ONE 2014;9(4): p.e93907; doi: 10.1371/journal.pone.0093907.
CrossRef - Jia B, Raphenya A. R, Alcock B, Waglechner N, Guo P, Tsang K. K, Lago B. A, Dave B. M, Pereira S, Sharma A. N, Doshi S, Courtot M, Lo R, Williams L. E, Frye J. G, Elsayegh T, Sardar D, Westman E. L, Pawlowski A. C and McArthur A. G. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic acids research, 2017; 45(D1):D566–D573; doi: https://doi.org/10.1093/nar/gkw1004.
CrossRef - Webb B and Sali A. Comparative Protein Structure Modeling Using MODELLER. Current Protocols in Bioinformatics 2016;54(1); doi: https://doi.org/10.1002/cpbi.3.
CrossRef - Tian W, Chen C, Lei X, Zhao J and Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Research 2018; 46(W1):W363–W367; doi: 10.1093/nar/gky473.
CrossRef - Ajayi G, Olagunju J, Ademuyiwa O, Ademuyiwa O and Martins O. C. Gas chromatography-mass spectrometry analysis and phytochemical screening of ethanolic root extract of Plumbago zeylanica, Linn. Journal of Medicinal Plants Research (2011); 5(9):1756–1761.
- Chandra D and Prasad K. Phytochemicals of Acorus calamus (Sweet flag). ~ 277 ~ Journal of Medicinal Plants Studies 2017;5(5):277–281.
- Singh B, Singh J. P, Kaur A and Singh N. Phenolic compounds as beneficial phytochemicals in pomegranate (Punica granatum L.) peel: A review. Food Chemistry 2018; 261:75–86; doi: 10.1016/j.foodchem.2018.04.039.
CrossRef - Sinha S, Sharma A, Reddy P. H, Rathi B, Prasad N. V. S. R. K and Vashishtha A. Evaluation of phytochemical and pharmacological aspects of Holarrhena antidysenterica (Wall.): A comprehensive review. Journal of Pharmacy Research 2013:6(4):488–492; doi: 10.1016/j.jopr.2013.04.004.
CrossRef - Azab S. S, Abdel-Daim M and Eldahshan O. A. Phytochemical, cytotoxic, hepatoprotective and antioxidant properties of Delonix regia leaves extract. Medicinal Chemistry Research 2013; 22(9):4269–4277; doi: 10.1007/s00044-012-0420-4.
CrossRef - Nanmeni G, Tedonkeu A. T, Fankam A. G, Mbaveng A. T, Wamba B. E. N, Nayim P, Bitchagno G. T. M, Nzogong R. T, Awouafack M. D, Tene M, Beng V. P and Kuete V. An Efflux Pumps Inhibitor Significantly Improved the Antibacterial Activity of Botanicals from Plectranthus glandulosustowards MDR Phenotypes. ScientificWorldJournal 2021;2021: 5597524; doi: 10.1155/2021/5597524.
CrossRef - Lunga P. K, Tamokou J. D, Fodouop S. P. C, Kuiate J, Tchoumboue J and Gatsing D. Antityphoid and radical scavenging properties of the methanol extracts and compounds from the aerial part of Paullinia pinnata. SpringerPlus2014; 3:302; doi: https://doi.org/10.1186/2193-1801-3-302.
CrossRef - Voukeng I. K, Beng V. P and Kuete V. Antibacterial activity of six medicinal Cameroonian plants against Gram-positive and Gram-negative multidrug resistant phenotypes. BMC Complement Altern Med 2016;16(1):388; doi: 10.1186/s12906-016-1371-y.
CrossRef - Seukep A. J, Kuete V, Nahar L, Sarker S. D and Guo M. Plant-derived secondary metabolites as the main source of efflux pump inhibitors and methods for identification. J Pharm Anal 2020; 10(4):277–290; doi: 10.1016/j.jpha.2019.11.002.
CrossRef - Tsopmejio J. P, Momeni J, Tsopjio F. N, Tedjon V. S, Mkounga P, Shehla N, Choudhary M. I and Nkengfack A. E. Bioactive secondary metabolites from Plectranthus glandulosus Hook. (Lamiaceae). Phytochemistry Letters 2019; 30: 133-137; doi: 10.1016/j.phytol.2019.01.029.
CrossRef
- Nyegue M. A, Afagnigni A. D, Ndam Y. N, Djova S. V, Fonkoua M. C and Etoa F. Toxicity and Activity of Ethanolic Leaf Extract of Paullinia pinnataLinn (Sapindaceae) in Shigella flexneri–Induced Diarrhea in Wistar Rats. J Evid Based Integr Med. 2020; 25:2515690X19900883; doi: 10.1177/2515690X19900883.
CrossRef - Bekro Y. A. Biological activities and phytochemical composition of the methanol extract of Paullinia pinnata Linn. International Journal of Current Research 2019;11(10):7728-7733; doi: 10.24941/ijcr.36903.10.2019.
- Mbaveng A. T, Tchana M. E. S, Fankam A. G, Nkwengoua E. T, Seukep J. A, Tchouani F. K, Nyassé B and Kuete V. Activities of selected medicinal plants against multi-drug resistant Gram-negative bacteria in Cameroon. African Health Sciences 2014; 14(1); doi: 10.4314/ahs.v14i1.25.
CrossRef - Lill M. A and Danielson M. L. Computer-aided drug design platform using PyMOL. Journal of Computer-Aided Molecular Design 2010;25(1):3–19; doi: 10.1007/s10822-010-9395-8.
CrossRef - Trott O and Olson A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010; 31(2): 455–461; doi: 10.1002/jcc.21334.
CrossRef - Salentin S, Schreiber S, Haupt V. J and Adasme M. S. PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Research 2015; 43(W1):W443–W447; doi: 10.1093/nar/gkv315.
CrossRef
- Fiser A. Template-Based Protein Structure Modeling. Methods Mol Biol. 2010; 673:73–94; doi: 10.1007/978-1-60761-842-3_6.
CrossRef
- Liao J, Wang Q, Wu F and Huang Z. In Silico Methods for Identification of Potential Active Sites of Therapeutic Targets. Molecules 2022; 27(20):7103; doi: 10.3390/molecules27207103.
CrossRef
- Takatsuka Y, Chen C and Nikaido H. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci U S A. 2010; 107(15):6559–6565; doi: 10.1073/pnas.1001460107.
CrossRef
- Lipinski C. A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 2004; 1(4):337-41; doi: 10.1016/j.ddtec.2004.11.007.
CrossRef - Veber D. F, Johnson S. R, Cheng H, Smith B. R, Ward K. W and Kopple K. D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem., 2002;45(12):2615-23; doi: 10.1021/jm020017n.
CrossRef - Ghose A. K, Viswanadhan V. N and Wendoloski J. J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases., J. Comb. Chem., 1999:1(55); doi: 10.1021/cc9800071.
CrossRef - Mierziak J, Kostyn K and Kulma A. Flavonoids as Important Molecules of Plant Interactions with the Environment. Molecules 2014; 19(10):16240–16265; doi: https://doi.org/10.3390/molecules191016240.
CrossRef - Aboulaghras S, Sahib N, Bakrim S, Benali T, Charfi S, Guaouguaou F, Omari N. E, Gallo M, Montesano D, Zengin G, Taghzouti K and Bouyahya A. Health Benefits and Pharmacological Aspects of Chrysoeriol. Pharmaceuticals (Basel). 2022; 15(8): 973; doi: 10.3390/ph15080973.
CrossRef - Huang D, Cheng J, Mao J, Ma S, Du Z, Chen W, Zhang F and Sun L. The LC-MS/MS-Based Measurement of Isopimaric Acid in Rat Plasma and Application of Pharmacokinetics. Biomed Res Int. 2021; 2021: 2310422; doi: 10.1155/2021/2310422.
CrossRef - Gupta S, Buttar H. S, Kaur G and Tuli H. S. Baicalein: promising therapeutic applications with special reference to published patents. PHARMACEUTICAL PATENT ANALYST 2022; 11(1); DOI: 10.4155/PPA-2021-0027.
CrossRef - Chen J, Li Z, Chen A. Y, Ye X., Luo H, Rankin G. O and Chen Y. C. Inhibitory effect of baicalin and baicalein on ovarian cancer cells. International Journal of Molecular Sciences 2013; 14(3):6012–6025; doi: 10.3390/ijms14036012.
CrossRef - Li J, Zhang H, Ning J, Sajid A, Cheng G, Yuan Z and Hao H. The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrobial Resistance and Infection Control 2019; 8:44; doi: 10.1186/s13756-019-0489-3.
CrossRef - Bilal S, Iqbal H, Anjum F and Mir A. Prediction of 3D structure of P2RY5 gene and its mutants via comparative homology modelling. Journal of Computational Biology and Bioinformatics Research 2009; 1(1):011-016.
- Daina A, Michielin O and Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017; 7: 42717; doi: 10.1038/srep42717.
CrossRef - Testa B and Krämer S. D. The biochemistry of drug metabolism–an introduction: Part 2. Redox reactions and their enzymes. Chemistry & biodiversity 2007; 4(3):257-405; doi: https://doi.org/10.1002/cbdv.200790032.
CrossRef - Di L, Kerns E. H. Drug-Like Properties: Concepts, structure design and methods from ADME to toxicity optimization (Second Edition). Academic Press: USA 2016; pp141-159.
- Pires D. E. V, Blundell T. L, Ascher D. B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. Journal of Medicinal Chemistry 2015; 58(9):4066–4072; doi: 10.1021/acs.jmedchem.5b00104.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





