Manuscript accepted on : 10 October 2011
Published online on: --
Naif Abdullah Al-Harbi
Department of Botany and Microbiology, Addiriyah Chair for Environmental Studies College of Science, King Saud University, B.O. Box 2455 Riyadh 11451, Saudi Arabia.
Corresponding Author E-mail: naalharbi@ksu.edu.sa
ABSTRACT: Production of protease enzyme from soil newly isolated Bacillus sp. (NASK P6) was investigated. The proteolytic activity was observed when the bacteria were grown in the minimal medium containing sucrose, K2HPO4, MgSO4, KCl2 and FeSO4 in shake flask culture condition. 16S rRNA phylogenetic analysis revealed that isolate NASK-P6 closely related to B. subtilis. Different inoculums size such as 1, 2, 3, 4, 5 and 10% in the same medium and further selected inoculums size with various carbon and nitrogen sources were used for production. The protease productivity was determined with various time interval i.e., 24, 48 and 72 hrs with 100 ml medium. The maximal productivity obtained (6319 U ml-1) in the 1% inoculums size at 72 hours, will be used to select best carbon, nitrogen and raw waste substrates. The maximum enzyme yield obtained in glucose (10564 U/ml/72 hrs), yeast extract (13777 U ml-1) and dhore dhal washing (19760 U ml-1) as most selective carbon, nitrogen and raw waste substrates respectively. Further the crude enzyme was subjected to various physical factors like temperature, pH and solvent stability analysed. The enzyme was highly active in alkaline pH (9.0), high temperature (55o C) and also active in acetone exposure.
KEYWORDS: Proteases; Bacillus; Thermo stable; Enzyme
Download this article as:| Copy the following to cite this article: Al-Harbi N. A. Production and Physical Characterization of Thermo-stable and Organic Solvents-tolerant Protease from Mesophilic Bacillus sp. NASK P6. Biosci Biotech Res Asia 2012;9(1) |
| Copy the following to cite this URL: Al-Harbi N. A. Production and Physical Characterization of Thermo-stable and Organic Solvents-tolerant Protease from Mesophilic Bacillus sp. NASK P6. Biosci Biotech Res Asia 2012;9(1). Available from: https://www.biotech-asia.org/?p=9497 |
Introduction
Enzymes are delicate protein molecules necessary for life. Proteases are essential constituents of all forms of life on earth, including prokaryotes, fungi, plants and animals. They can be cultured in large quantities in a relatively short time by established methods of fermentation and they also produce an abundant, regular supply of the desired product. Microorganisms account for a two third share of commercial protease production in the world (Kumar and Takagi 1999). Protease are the single class of enzymes which occupy a pivotal position due to their wide applications in detergent, pharmaceutical, photography, leather, food and agricultural waste processing industries (Pastor et al, 2001;Ward, 1985). An important biotechnological application of protease is in bioremediation processes (Gupta et al. 2002). Among the various proteases, bacterial proteases are the most significant, compared with animal and fungal proteases and among bacteria, Bacillus sp. are specific producers of extracellular proteases. Bacilus subtilis is one of the most widely used bacteria for the production of specific chemicals and industrial enzymes and also a major source of amylase and protease enzymes. Now a days new enzyme are constantly being sought. Established industrial enzymes are being used in a variety of ways, in reaction alien to their natural functions. Earlier reports have shown that enzymes can be active in organic solvents which have greatly expanded their potential for use in syntheses of useful products. As catalysts in the presence of organic solvents, enzymes must be stable. The reaction of these enzymes can be enhanced by modifying the enzyme so as to make it more compatible to be use in synthesis in organic solvent. Methods (physical and chemical) have been developed for stabilizing enzymes in the extracellular organic solvent tolerant protease producer has been successfully isolated from various environments (Geok et al, 2003).
Materials and Methods
Bacterial strain
Bacillus sp. (NASK P6) was isolated from rhizosphere soil of dates palm tree in and around Riyadh. The collected soil samples diluted and spreaded over Skim-milk agar plates containing (g l-1): peptone 5 yeast extract 3, bacteriological agar 12 and skim-milk 250 ml, pH 10.0. Plates were incubated 24–48 h at 37 ◦C. A clear zone of skim-milk hydrolysis gave an indication of protease-producing strains. Individual colonies were purified through repeated streaking on fresh agar plates. Among the 35 isolates, showing different proteolytic activities, isolate NASK P6 was selected. The strain was identified as Bacillus sp.
16S rRNA amplification and phylogenetic analyses
DNA from NASK P6 isolate was extracted using the Illustra bacteria genomicPrep Mini Spin Kit (GE Healthcare UK Limited, UK). The 16S rRNA gene was amplified by PCR using the universal bacterial primers 8F and 1492R as described by Duckworth et al. (1996). Sequencing was performed on both strands of PCR amplified fragments using the dideoxy chain-termination method by the commercial services of MWG Biotech Pvt. Ltd. (Bangalore, India). 16S rRNA gene sequence similarity was carried out against the NCBI database using BLAST (Altschul et al., 1990). Sequences showing a relevant degree of similarity were imported into CLUSTAL W program (Thompson et al., 1994), and it was aligned and corrected manually. Phylogenetic analyses were done according to two different methods: maximum likelihood (ML) and maximum-parsimony (MP) with MEGA 4.0 (Tamura et al., 2007). The results were evaluated with 1,000-replication bootstrap analysis. Strains numbers and their corresponding accession numbers are indicated on the phylogenetic trees (Fig.1)
 |
Figure 1: Maximum-parsimonious phylogenetic tree derived from 16S rRNA gene sequence data, showing the phylogenetic affiliation of the proteolytic isolates.
|
Organism and Inoculum
Bacillus sp. NASK p6 regularly cultivated in nutrient broth medium for maintenance and, inoculum prepared with a loop full of culture was inoculated in 50ml of sterile nutrient broth and incubated at 37°C for 24 hrs. Further the culture was used as a source of inoculum.
Production medium
Control medium
The protease enzyme production was carried out with the modified Czapek Dox minimal medium containing Sucrose, K2HPO4, MgSO4, KCl2 and FeSO4.
Control medium and inoculums size
Control medium was inoculated with 1%, 2%, 3%, 4%, 5% and 10% inoculums size given in 100ml of sterile medium and incubated for 37°C for 24 , 48 and 72 hrs incubation with shake culture 120rpm.
Production media with different Carbon and Nitrogen sources
Along with control medium various carbon sources i.e. glucose, lactose, galactose, gelatin, starch and by varying nitrogen sources such as malt extract, peptone, and yeast extract were used. The standard control medium also combined with waste products for production of enzyme i.e., Rice, broken wheat washing as carbon source and Dhore dhal, Mung dhal, Bengal gram, and black gram washings as nitrogen source. The production medium was inoculated with 1% inoculums size further incubated in shake culture with 120rpm at 37°C for 24, 48 and 72hrs.
Protease assay
Protease activity was measured by the method described by Kembhavi et al. 1993, using casein as a substrate. One unit of protease activity was defined as the amount of enzyme required to liberate 1mg of tyrosine per minute under the used experimental conditions. Protease activities represent the mean of at least two determinations carried out in triplicate.
Effect of pH on protease activity and stability
The optimum pH of the crude enzyme was studied over a pH range of 4.0–12.0 at 37 ◦C using casein 1% (w/v) as a substrate. The effect of pH on proteolytic stability was examined by incubating the enzyme in buffers of different pH values in the range of pH 5.0–13.0 for 1 h at 25 ◦C. Aliquots were withdrawn at the desired time intervals to test the remaining caseinolytic activity at pH 10.0 and 70 ◦C. The following buffer systems were used for this study;
pH range (4-5) – Glycine – HCL buffer
pH range (6 – 9) – Phosphate Buffer
pH range (10 – 11) – Glycine – Sodium Hydroxide Buffer.
Effect of temperature on protease activity
The effect of temperature on protease activity was studied from 10◦C, 25◦C, 37◦C, 45◦C, 55◦C and 65◦C. Thermo-stability was examined by incubating the crude enzyme for 120 min. Aliquots were withdrawn at desired time intervals to test the remaining activity at standard conditions using casein as a substrate
Organic solvent stability assay
The effect of various organic solvents on the stability of the crude enzyme was studied. The crude enzyme was incubated along with 90% v/v of various organic solvents (acetone, ethanol, methanol and petroleum Ether) at 37 ◦C with shaking (150 rpm) for 15 min, 30min and subsequent incubation for 6 days. Aliquots were withdrawn at desired time intervals to test the remaining activity at standard conditions. The activity of the enzyme measured in absence of any organic solvent was taken as control.
Results and Discussion
Isolation, characterization and phylogenetic analyses of extracellular protease-producing bacteria
Totally 35 isolates were isolated from 10 different soils of dates palm tree in Riyadh city. Among the 35 isolates, six isolates showed porteolytic enzyme, the isolate NASK-P6 was selected for further studies based on the maximal protease enzyme production. The isolate NASK-P6 was aerobic, endospore-forming, Gram-positive and rod-shaped bacterium. Isolate NASK-P6 identified as Bacillus sp. based on their biochemical and physiological characteristics. The 16S rDNA sequences were analyzed and determine the BLAST studies cited in the GenBank matched the isolate with the highest homology. Sequence data were aligned to construct phylogenetic trees using the MEGA 4.0, and are shown in Fig. 1. The phylogenetic position of isolate Bacillus sp. NASK-P6 was compared with Bacillus species obtained from GenBank in a phylogenetic tree. Isolate NASK-P6 was found to be closest to Bacillus tequilensis, B. axarquiensis, B. amyloliquefaciens and B. subtilis with 100% of similarity matrix and maximum identity.
On the other hand, the results of the phylogenetic analyses, based on the 16S rRNA gene, the overall topology of the tree based on analysis, the isolate NASK-P6 was more closely related to B. subtilis ASLC18B (HM585072) compare than other Bacillus species. Nevertheless, this group not confirmed by strong bootstrap support (58%).
Screening of appropriate inoculums size for protease
The result of our study showed that production of protease increased with inoculums size at 24 hrs in 2 % to 10 % rather than 1% (Fig. 2). The production increased linearly with increase in incubation time till 72 hrs in 1% but further incubation showed reduction in enzyme production for 2 % to 10 % inoculum sizes (Fig. 2). Maximum protease production was observed in 1 % of inoculum (6319 U ml-1) at 72 hrs in production medium. Bacterial DNA content increased with increase in incubation time similar to protease production suggesting that enzyme production was growth associated in nature. These results are in accordance with observations made by Gessesse (1997), Yeoman and Edwards (1994) and Johnvesly et al. (2002).
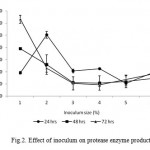 |
Figure 2: Effect of inoculum on protease enzyme production.
|
Selection of most suitable carbon and nitrogen sources by one-variable-at-a-time approach
The Bacillus sp. produced maximum protease in the presence of glucose (10564 U ml−1) followed by gelatin (8795U ml−1), lactose (5933 U ml−1), galactose (4004 U ml−1), fructose (3206 U ml−1) and starch (2866 U ml -1) at 72 hrs incubation time as shown in Fig. 3. The readily assailable simple carbon sources, viz. fructose, galactose, lactose supported poor protease production. Similarly, complex nitrogen sources, viz. peptone (10333 Uml−1), yeast extract (13777 U ml−1) and malt extract (8524 U ml−1) and yeast extract (13777 U ml−1) were observed to induce high protease production while the production was steadily increased incubation time increases perpetually (Fig 4). Thus, glucose and yeast extract were enhanced the maximum productivity of isolate for this study.
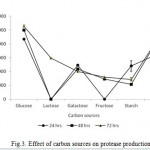 |
Figure 3: Effect of carbon sources on protease production.
|
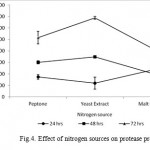 |
Figure 4: Effect of nitrogen sources on protease production.
|
The production of extracellular protease by Bacillus sp. is 80 fold higher in the presence of simple carbon and nitrogen sources, viz. glucose and yeast extract as compared to simple sugars, viz. fructose, galactose or nitrogen sources, viz. peptone, malt extract, etc. There are similar reports of enhanced protease production in the presence of complex carbon (Gupta et al, 2002; Hubner et al, 1993) and other nitrogen sources (Gupta et al, 2002; Puri et al, 2002; Joo et al, 2002).
Evaluation of different agro-industrial material for alkaline protease production
The selection of ideal gray kitchen waste water for enzyme production in a solid-state fermentation process depends upon several factors, mainly related with cost and availability of the substrate material, and thus may involve screening of several agro-industrial residues (Pandey et al., 2000). The results in the present study indicated that protease production pattern varied with the type of gray kitchen waste water. This could be attributed to solid materials dual role-supply of nutrients to the microbial culture growing and anchorage for the growing cells.
Maximum enzyme production (19760 U ml-1) was observed with dhore dhal wash, while minimum protease production (6522 U ml-1) was noticed with rice wash as substrate/support material in 72 hrs incubation (Fig 5). In this present study showed the growth and enzyme production were eventually increased in all the gray kitchen waste water at different time intermittence. These results are in accordance with the observations made with alkalophilic and thermophilic Bacillus sp. JB-99 (Johnvesly et al., 2002) and other reported strains (Gessesse, 1997). Evaluation of protease production values of this strain with different literature reported organisms indicated that this strain could be a potential source after optimization and scale-up.
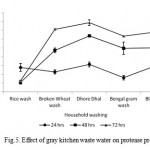 |
Figure 5: Effect of gray kitchen waste water on protease production.
|
The protease production by Bacillus sp. NASK P6 was found to be inducible. There was pronounced induction in the presence of nitrogen sources say, yeast extract and peptone. Microbial protease production has been found to vary from being constitutive to partially inducible in nature (Gupta et al., 2002; Puri et al., 2002; Gusek et al., 1988). Also, protease production was under growth independent repression in the presence of high concentrations of yeast extract. There are several reports on the repressive role of nitrogen sources and other waste materials.
Effect of physical factors and organic solvent-stability of crude enzyme
Protease production by microbial strains strongly depends on the extra-cellular pH because culture pH strongly influences many enzymatic processes and transport of various components across the cell membranes, which in turn support the cell growth and product production (Ellaiah et al., 2002). The protease enzyme subjected to various pH range from 4 – 11. Results showed that enzyme synthesis was increased with increase of medium pH towards alkaline range from neutrality, maximum (15779 U ml-1) being at pH 10.0 (Fig. 6) and then remained more or less constant in the pH range 9.0–11.0 (results not shown). This suggested that the bacterial strain was alkalophilic in nature. The protease activities were assayed at different temperatures ranging from 10 to 90°C at a constant pH of 7.0 (Fig.7). Enzyme activity increased with temperature within the range of 45 to 65°C. A reduction in enzyme activity was observed at values above 25°C. The optimum temperature of this protease was 55°C. The maximum activity was observed at 55oC (11890 U ml-1).
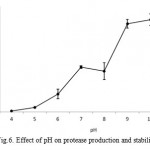 |
Figure 6: Effect of pH on protease production and stability.
|
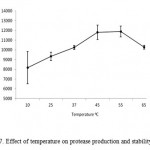 |
Figure 7: Effect of temperature on protease production and stability.
|
Enzymes are usually inactivated or denaturated in the presence of organic solvents (Ghorbel et al., 2003). The effects of different organic solvents viz., acetone, ethanol, methanol, petroleum ether on protease activity were studied. The relative activity, which remained after 15 min, 30 min and 6 days of incubation in 90% (v/v) of organic solvent at 37oC, is shown in Fig. 8. The activity of the enzyme, without any solvent (control), was taken as 100%. Bacillus sp. NASK P6 protease showed a remarkable stability in the presence of all of the solvents, as shown in Figure 2. The maximum activities were present in acetone (13970 U ml-1) than other organic solvents used. The activities of alkaliphilic protease from Bacillus subtilis TKU07, were 65% and 90% in the presence of only 20% (v/v) of butanol and toluene (Wang andYeh, 2006). Gupta and Khare (2007) reported that crude P. aeruginosa PseA protease showed a remarkable stability in the presence of most solvents, having the logarithm of the partition coefficient (log P) above 2.0, but was less stable in the presence of hydrophilic solvents.
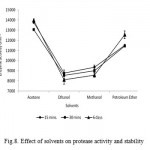 |
Figure 8: Effect of solvents on protease activity and stability.
|
Conclusion
The protease production by soil isolated Bacillus sp. NASK P6 strain under solid-state fermentation was influenced by physiological and ingredients nature of dhore dhal (gray kitchen waste water) and associated with growth of the microbial strain. Simplicity of the medium components was found to be important to achieve maximum enzyme production yields. Dhore dhal, as nutrient source, was insufficient medium for production of enzyme. Enhanced protease production yields would be obtained by supplementation of glucose and yeast extract to the solid medium.
References
- Altschul, S.F, Gish, W., Miller, M., Myers, E.W., Lipman, D.J., 1990. Basic local alignment search tool. J Mol Biol 215:403–410.
- Duckworth, A. W., Grant, W. D., Jones, B. E., van Steenbergen, R., 1996. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol Ecol 19, 181–191.
- Ellaiah, P., Srinivasulu, B., Adinarayana, K., 2002. A review on microbial alkaline proteases. J. Sci. Ind. Res. 61, 690–704.
- Geok LP, Razak CNA, Rahman RNZA, Basri M, Salleh AB. 2003. Isolation and screening of an extracellular organic solvent tolerant protease producer. Biochem Eng J. 13:1–5.
- Gessesse, A., 1997. The use of nug meal as a low-cost substrate for the production of alkaline protease by the alkaliphilic Bacillus sp. AR- 009 and some properties of the enzyme. Bioresour. Technol. 62, 59– 61.
- Ghorbel B, Kamoun AS, Nasri M. 2003. Stability studies of protease from Bacillus cereus BG1. Enzyme Microb Tech 32:513-518.
- Gupta A, Khare SK. 2007. Enhanced production and characterization of a solvent stable protease from solvent tolerant Pseudomonas aeruginosa PseA. Enzyme Microb Tech 42:11-16.
- Gupta R, Beg QK, Khan S, Chauhan B. 2002An overview on fermentation downstream processing and properties of microbial alkaline proteases. Appl Microbiol Biotechnol. 60:381–95.
- Gupta R, Beg QK, Lorenz P. 2002. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 59 (1): 15-32.
- Gusek TW, Wilson DB, Kinselle JE. Influences of carbon source on production of heat stable protease from Thermomonospora fusca YX. Appl Microbiol Biotechnol 1988;28:80–4.
- Hubner U, Block U, Schugerl K. Production of alkaline serine protease subtilisin Carlsberg by Bacillus licheniformis on complex medium in a stirred tank reactor. Appl Microbiol Biotechnol 1993;40:182–8.
- Johnvesly, B., Manjunath, B.R., Naik, G.R., 2002. Pigeon pea waste as a novel, inexpensive, substrate for production of a thermostable alkaline protease from thermoalkalophilic Bacillus sp. JB-99. Bioresour. Technol. 82, 61–64.
- Joo HS, Kumar CG, Park GC, Kim KM, Paik SR, Chang CS. Optimization of the production of an extracellular alkaline protease from Bacillus horikoshi. Process Biochem 2002;38:155–9.
- Kembhavi AA, Kulharni A, Pant AA. 1993. Salt-tolerant and thermostable alkaline protease from Bacillus subtilis NCIM No. 64. Appl Biochem Biotechnol. 38:83–92.
- Kumar CG, Takagi H. 1999. Microbial alkaline proteases: from a bioindustrial viewpoint. Biotechnol Adv 17:561–594
- Pastor MD, Lorda GS, Balatti A. 2001. Protease obtention using Bacillus subtilis 3411 and amaranth seed meal medium at different aeration rates. Braz. J. Microbiol. 32: 1-8.
- Puri, S., Beg, Q.K., Gupta, R.G., 2002. Optimization of alkaline protease production from Bacillus sp. using response surface methodology. Curr. Microbiol. 44:286–90.
- Tamura, K., Dudley, J., Nei, M., Kumar, S., 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24:1596-1599.
- Thompson, J.D., Higgins, D.G., Gibson, T.J., 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680
- Wang SL, Yeh PY. 2006. Production of a surfactant- and solvent-stable alkaliphilic protease by bioconversion of shrimp shell wastes fermented by Bacillus subtilis TKU07. Process biochem 41:1545-1552.
- Ward OP. 1985. Proteolytic enzymes. In: Blanch, h.W., Drew, S., Wang, D.I., eds; comprehensive Biotechnology. Vol. 3. Oxford U.K., Pergamon Press. 789-818.
- Yeoman, K.H., Edwards, C., 1994. Protease production by Streptomyces thermovulgaris grown on rapemeal-derived media. J. Appl. Bacteriol. 77, 264–270.

This work is licensed under a Creative Commons Attribution 4.0 International License.





