How to Cite | Publication History | PlumX Article Matrix
Decolourization of Azo-Dyes by Bacterial Consortium
M. N. Abubacker¹* and A. Ayesha²
¹Department of Biotechnology, National College, Tiruchirappalli - 620 001 India
²Department of Civil Engineering, Kongu Engineering College, Perundurai, Erode - 638 052 India.
Corresponding Author E-mail: abubacker_nct@yahoo.com
ABSTRACT: Azo-dye polluted soil samples were used for the isolation of bacterial species. Among the isolates Enterobacter aerogens, Proteus rettgeri, Pseudomonas fluorescens, and Staphylococcus aureus were the dominant bacterial species present and they are designated as indicator bacterial isolates. These indicator bacterial isolates are able to utilize the dye as nitrogen source and hence they are able to decolorize the azo-red and blue dyes. Decolourization was assayed colorimetrically at 540 nm for red and 580 nm for blue and percentage of decolourization was calculated. The optimum concentration for both the dyes was 100 ppm. The maximum decolourization of azo-red and azo-blue dye at the end of 96 hours of decolourization experiments were 90% and 80% respectively for consortium of indicator bacterial isolates. The individual bacterial isolates were less effective for decolourization. These bacterial consortium can be exploited as bioremediation agents to reduce dye pollutants.
KEYWORDS: Azo-red; Azo-blue dyes; Consortium; Decolourization; Bacterial isolates; Pollutant
Download this article as:| Copy the following to cite this article: Abubacker M. N, Ayesha A. Decolourization of Azo-Dyes by Bacterial Consortium. Biosci Biotech Res Asia 2011;8(2) |
| Copy the following to cite this URL: Abubacker M. N, Ayesha A. Decolourization of Azo-Dyes by Bacterial Consortium. Biosci Biotech Res Asia 2011;8(2). Available from: https://www.biotech-asia.org/?p=9559 |
Introduction
Water pollution control is at present one of the major thrust areas of scientific research. Colour removal in particular, has recently become an area of major scientific interest as indicator (Moosvi et al., 2005). Dyes are released into the environment through industrial effluents from three major sources such as textile, dye stuff manufacturing and paper industry. One of the most pressing environmental problems related to dye effluents is the improper disposal of waste water from dyeing industry (Rajeswari et al., 2011).
Dyes are chemical substances, which are used to colour textile fabrics. Dyes are synthetic, aromatic and water-soluble dispersible organic colorants, having potential application in various industries. Dyes include acidic, basic, azoic, chromic, diazoic dispersive, reactive sulphur and vat dyes. Approximately 1,00,000 commercial dyes are manufactured with an annual production of over 7´105 metric tons (Campos et al., 2001; Mohan et al., 2002).
In the azo dyes the aromatic moieties are linked together by azo (– N = N –) chromophores, and they are the largest class of dyes employed in dyeing and printing processes. The downstream processes, i.e., after printing, washing and finishing, generate large quantities of coloured waste water (Othman et al., 2011). Colour in any water is undesirable and is therefore considered a pollutant. Dyes are stable to light and oxidizing agents and are sometimes resistant to bio-oxidation. Partial degradation products of azo dyes are aromatic amines which have toxic effects. The strong colour of discharged dyes even at very small concentrations has a huge impact on the aquatic environment caused by its turbidity and high pollution strength (Gondaliya and Pathak, 2005).
Coloured industrial effluents from the dyeing industries represent major environmental problems. Dye waste water discharged from textile and dyestuff industries have to be treated. Colour can be removed from waste water by physico-chemical methods. These include adsorption, coagulation, chemical transformation, flocculation, oxidation, filtration, photocatalysis, ozonation and electrochemical methods. These methods are often laborious and expensive (Sathiyamoorthi et al., 2007). The success of biological process for colour removal from a given effluent depends in part on the utilization of microorganisms that effectively decolorize synthetic dyes of different chemical structures. Many bacteria, actinomycetes, yeast and fungi are able to decolorize dyes (Machado et al., 2006). Biodecolourization constitutes an attractive alternative to physico-chemical methods as a low cost, eco-friendly and publicly acceptable technology. The removal of the polluting dyes is a serious problem, particularly for small scale textile industries where working conditions and low economic status do not allow them to treat their wastewater before disposal and they have no choice other than to dump all effluents into the main stream of water sources. Microbial decolourization and degradation is an eco-friendly and cost-competitive alternative to chemical decomposition processes (Verma and Madamwar, 2003). At present, bioremediation relies on the pollutant degrading capacities of naturally occurring microbial consortia in which bacteria play a control role (O’Neill et al., 2000). The ability of microorganisms to carry out dye decolourization has received much attention as microbial decolourization of dyes is a cost effective method for removing them from the environment (Kalyani et al., 2008). Currently, extensive research is being focused on finding an optimal microbial biomass that would be as cheap as possible for the removal of contaminating dyes from large volume of polluted water (Youssef et al., 2008). In this study, an attempt was made to isolate bacterial consortium from dye waste contaminated soil and their decolourization efficiency was studied.
Material and Methods
Isolation of Bacteria
Soil samples were collected from dye effluent contaminated sites at Karur in aseptic condition. Soil suspension was prepared by mixing 10 gm of soil in 100 ml of sterile distilled water. The samples were serially diluted and 0.1 ml of the suspension was spread plated on Nutrient Agar (Himedia) plates in sterile condition. The plates were incubated at 32° C ± 1° C for 2 days. The bacterial colonies grown on the plates were further sub-cultured and pure colonies were isolated and stored on Nutrient Agar slants for further investigation.
Screening of Dye Degradation
The isolated pure bacterial colonies were screened for dye decolourization by incorporating 0.1 mg / 100 ml of azo-red and azo-blue in Nutrient agar medium. Then the plates were incubated at 32° ± 1° C for 2 days to observe the clear zone formed due to the degradation of the dye. The bacteria which showed distinct zone of clearance were selected, identified and used for further study.
Decolourisation of Dye
The isolated bacterial cultures were incorporated in nitrogen limited medium (Glucose 15 g, malt extract 0.4 gm, manganese chloride 0.3 g, ferric sulphate 0.4 g, magnesium sulphate 0.04 g, pH 6.0, distilled water 1000 ml) and amended with dye at a concentration of 100 to 500 ppm. The pH of the dye solution was 6.5 to 6.8. The medium with dye solutions were sterilized and used for decolourization studies. A loop of pure culture was inoculated into the medium at 32° ± 1° C and decolourization was observed for 96 hours. The content was assessed for decolourization by measuring absorbance at 540 nm (Red), 580 nm (Blue) using UV-visible spectrophotometer at the end of 24, 48, 72 and 96 hours. Decolourization was expressed in terms of percentage and calculated using the following formula used by Olukanni et al. (2006).
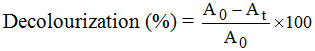
where A0 = Absorbance of the blank (dye solution)
At = Absorbance of the treated dyes solution at specific time
Results and Discussion
Dyes are aromatic compounds. They are degraded by bacteria during metabolism. It is important to ensure constant and complete removal of these dyes and their metabolites from the environment to prevent setting up of bioaccumulation.
Twelve bacterial cultures were isolated from the dye contaminated soil by spread-plate technique. Among them Enterobacter aerogenes, Proteus rettgeri, Pseudomonas fluorescens, and Staphylococcus aureus showed maximum zone of clearance in azo-red and azo-blue dye amended Nutrient Agar medium. Hence these bacteria were identified as indicator bacteria of dye degradation and were selected for this study. Maximum 70 per cent of decolourization at 96 h was shown by P. fluorescens for 100 ppm concentration of azo-red dye followed by 60, 55 and 50 per cent respectively for E. aerogenes, P. rettgeri and S. aureus. For azo blue dye maximum 65 per cent of decolourization at 96 h was shown by P. fluorescens followed by 50 per cent by E. aerogenes and P. rettgeri whereas 45 per cent by S. aureus. It was observed that a consortium of all these bacteria showed better result than the individual bacterial species and were able to decolorize 90% for azo-red dye and 80% for azo-blue at 32° ± 1° C in 96 hours (Tables 1-3 and Fig. 1-3).
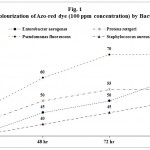 |
Figure 1
|
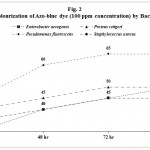 |
Figure 2
|
 |
Figure 3
|
The decolourization effect of different bacterial species is variable. It has been reported that the adsorption of chromatophores on the cell surface of the microorganisms results in the decolourization of dyes. The possible mechanism is biosorption which is dependent on functional groups in the dye molecule on bacterial biomass, which also plays a role in the biosorption of dyes (Fu and Viraraghavan, 2002). Rodriguez et al. (1999) reported that several industrial dyes were decolorized biocatalytically by extracellular enzymes of microbes. Chang and Lin (2000) reported that azo dyes are effectively decolorized by Pseudomonas luteola strain. Mabrouk and Yusef (2008) reported decolourization of fast red by Bacillus subtilis. Olukanni et al. (2006) reported decolourization of azo dyes by strain of Micrococcus. Nortemann et al. (1986) revealed that bacterial communities were able to degrade amino and hydroxyl-naphthalene-sulfonates in azo dye. Hong et al. (2000) reported that Bacillus pulmilus bacteria is involved in biosorption of 1,2,3,4-tetrachlorodibenzo-p-dioxin of azo dye. The present study revealed 100 ppm as optimum concentration, pH 5.8, temperature at 32° ± 1° C as optimum conditions for maximum decolourization by bacterial consortium for both azo-red and azo-blue dyes. These results are on par with the previous works of Chang and Lin (2000), McMullan et al. (2001), Olukanni et al. (2006) and Mabrouk and Yusef (2008).
Table 1: Decolourization of Azo-red dye (100 ppm concentration) by Bacteria.
| Bacteria | Percentage of Decolourization | |||
| 24 hr | 48 hr | 72 hr | 96 hr | |
| Enterobacter aerogenes | 35 | 45 | 50 | 60 |
| Proteus rettgeri | 40 | 50 | 55 | 55 |
| Pseudomonas fluorescens | 45 | 60 | 70 | 70 |
| Staphylococcus aureus | 35 | 40 | 45 | 50 |
Table 2: Decolourization of Azo-blue dye (100 ppm concentration) by Bacteria
| Bacteria | Percentage of Decolourization | |||
| 24 hr | 48 hr | 72 hr | 96 hr | |
| Enterobacter aerogenes | 30 | 40 | 45 | 50 |
| Proteus rettgeri | 40 | 45 | 50 | 50 |
| Pseudomonas fluorescens | 45 | 60 | 65 | 65 |
| Staphylococcus aureus | 35 | 40 | 45 | 45 |
Table 3: Decolourization of Azo-red and Azo-blue (100 ppm concentration) by Bacterial Consortium (Enterobacter aerogenes, Proteus rettgeri, Pseudomonas fluorescens, and Staphylococcus aureus)
| Dye | Percentage of Decolourization | |||
| 24 hr | 48 hr | 72 hr | 96 hr | |
| Azo-red dye | 45 | 60 | 70 | 90 |
| Azo-blue dye | 40 | 50 | 70 | 80 |
Conclusion
The present study is thus an effort to develop a bacterial-based treatment system for the cleaning up of dye industry effluents and for bioremediation of dye contaminated soil using bacterial-based consortium.
Acknowledgements
We wish to thank DST-FIST Government of India for Instrumentation facilities provided to the Department of Botany, National College, Tiruchirappalli. The authors thank Sri. K. Regunathan, Secretary and Dr. K. Anburasu, Principal, National College for their encouragement. We also thank Dr. S. Kuppuswami, Principal; Prof. S. Krishnamoorthi, Head; and Prof. N. Rajkumar, Department of Civil Engineering, Kongu Engineering College, Perundurai, Erode for their encouragement.
References
- Campos, R., Kandelauer, A, Robra, K. H., Artur Cavalo, Paulo and Gubitz, G. M., Indigo degradation with purified laccases from Trametes hirsuta and Sclerotium roltsie. J. Biotechnol. 8: 131-139 (2001).
- Chang, J. S. and Lin, Y. C., Fed-batch bioreactor strategies for microbial decolorization of azo dye using a Pseudomonas luteola strain. Biotechnol Prog. 16: 979 (2000).
- Fu, Y. and Viraraghavan, T., Dye biosorption sites in A. niger. Bioresource Technology. 82: 139-145 (2002).
- Gondaliya, S. B. and Pathak, S. J., Isolation of textile dyes decolourizing microbes and their optimization of culture conditions. Asian Journal of Microbiology, Biotechnology and Environmental Sciences. 7: 545-553 (2005).
- Hong, H., Hwang, S. and Chang, Y., Biosorption of 1,2,3,4-tetra-chloro-dibenzo-p-dioxin and polychlorinated dibenzofurans by Bacillus pulmilus. Water Res. 34: 349 (2000).
- Kalyani, D. C., Patil, P. S., Jadhav, J. P. and Govindwar, S. P., Biodegradation of reactive textile dye Red BLI by an isolated bacterium Pseudomonas sp. SUK1. Bioresour. Technol. 99: 4635-4641 (2008).
- Mabrouk, M. E. M. and Yusef, H. H., Decolourization of Fast Red by Bacillus subtilis. H. M. J. Appl. Sci. 4(3): 262-269 (2008).
- Machado, K. M. G., Compari, L. C. A., Morais, R. O., Luiz, H., Rosa Mercia H. Santos, Biodegradation of reactive textile dyes by Basidiomycetous fungi from Brazilian ecosystems. Brazilian J. Microbiol. 37: 481-487 (2006).
- McMullan, G., Meechan, G., Conneely, A., Kirby, N., Robinson, T., Banat, I., Marchant, R. and Smyth, W. F., Microbial decolourisation and degradation of textile dyes. Applied Microbiol Biotechnology. 56: 81-85 (2001).
- Mohan, S. V., Rao, C. N., Prasad, K. K., Karthikeyan, J., Treatment of simulated reactive yellow 22 (azo) dye effluents of Spirogyra sp. Waste Manage. 22: 575-582 (2002).
- Moosvi, S., Keharia, H. and Madamma, D., Decolourization of textile dye reactive violet 5 by a newly isolated bacterial consortium RVM 11.1. World J. Microbiol. Biotechnol. 21: 667-672 (2005).
- Nortemann, B., Baumgarten, J. Rast, H. G. and Knackmuss, H. J., Bacterial communities degrading amino- and hydroxynaphthalenesulfonates. Appl. Environ. Microbial. 52: 1195 (1986).
- Olukanni, O. D., Osuntoki, A. A. and Gbenle, G. O., Textile effluent biodegradation potentials of textile effluent adapted and non-adapted bacteria. African Journal of Biotechnology. 5: 1980-1984 (2006).
- O’Neill, C., Lopez, A., Esteves, S., Hawkes, F. R., Hawkes, D. L. and Wilcox, S., Azo-dye degradation in an anaerobic-aerobic treatment system operating on simulated textile effluent. Applied Microbiol. Biotechnol. 53: 249-254 (2000).
- Othman, N., Mill, N. and Wong, Y. M., Liquid-liquid extraction of black B dye from liquid waste solution using tridodecylamine. J. Environ. Sci. Technol. 4: 324-331 (2011).
- Rajeshwari, K., Subashkumar, R. and Vijayaraman, K., Biodegradation of Mixed Textile Dyes by Bacterial Strains Isolated from Dyewaste Effluent. Res. J. Environ. Toxicol. 5: 97-107 (2011).
- Rodriguez, E., Pickard, M. A. and Duhatt, R. V., Industrial decolourization by lactase from ligninolytic fungi. Current Microbiology. 38: 27-32 (1999).
- Sathiyamoorthi, P., Periyar Selvam, S., Sasikalaveni, A., Murugesan, K. and Kalaichelvan, Decolourization of textile dyes and their effluents using white rot fungi. African J. Biotechnol. 6: 424-429 (2007).
- Verma, P. and Madamwar, D., Decolourization of synthetic dyes by a newly isolated strain of Serratia marcescens. World J. Microbiol. Biotechnol. 19: 615-618 (2003).
- Youssef, A. S., El-Sherif, F. M. and El-Assar, A. S., Studies on the decolourization of malachite green by the local isolate Acremonium kiliense. Biotechnology. 7: 213-223 (2008).

This work is licensed under a Creative Commons Attribution 4.0 International License.





