How to Cite | Publication History | PlumX Article Matrix
Mohammad Hossein Morowvat1, Navid Nezafat1, Younes Ghasemi1,2٭, Mohammad Hadi Zare1,2, Milad Mohkam2
1Pharmaceutical Sciences Research Center, Shiraz University of Medical Sciences,P.O. Box 71345-1583, Shiraz, Iran 2Department of Pharmaceutical Biotechnology, Faculty of Pharmacy, Shiraz University of Medical Sciences,P.O. Box 71468-64685, Shiraz, Iran
ABSTRACT: Probiotics are dietary supplements of live microorganisms which presence of them in gastrointestinal tract in sufficient quantities can be beneficial to host health. Amongst various bacterial strains, lactic acid bacteria (LAB) are the most common probiotic bacteria. The aims of this study were to isolate and identify potentially LAB probiotics from the fermented yoghurt products of townships and villages in Fars province, Iran. Thirty samples of traditional dairy products were collected from different regions of Fars province. Laboratory methods such as gram-staining, motility; and biochemical approaches including carbohydrate utilization and catalase activity were employed to isolate LAB and assess their relevant properties. In five final isolates, 16S rDNA genes were extracted, amplified through PCR and electrophoresed. These genes were then sequenced and compared with the library of the sequences of the NCBI databases. Five different isolates of Lactobacillus were identified and reported as the normal flora in traditional dairy products. These included L. acidophilus, L. delbrueckii subsp. bulgaricus, L. jensseni, L. cripatus and L. gasseri. Phylogenetic analysis revealed a genetic relationship between the five studied strains together and with some previously reported probiotic strains. The obtained strains are potentially probiotics and these isolates can be used in dairy product industries to achieve beneficial effects of probiotics.
KEYWORDS: probiotic properties; lactic acid bacteria; traditional dairy products; functional food; phylogenetics
Download this article as:| Copy the following to cite this article: Morowvat M. H, Nezafat N, Ghasemi Y, Zare M. H, Mohkam M. Probiotic Potential of Five Lactobacillus Strains Isolated from Traditional Persian Yoghurt in Fars Province, Iran: Viewing Through the Window of Phylogenetics. Biosci Biotech Res Asia 2015;12(2) |
The Probiotic term, that derived from two Greek words Pro (favor) and bios (life) (Jothi et al., 2012), is defined as live microorganisms which transit the gastrointestinal tract and in doing so, they benefit the health of host (Tannock et al., 2000). Lactobacilli are gram-positive microorganisms which belong to the general category of lactic acid bacteria (LAB) and potentially could be considered as probiotics. This genus consists of a genetically and physiologically diverse group of rod-shaped, non-spore forming, non-pigmented, catalase negative and microaerophilic to strictly anaerobic bacteria which have widespread use in fermented food production (Hassan and Frank, 2001; Coeuret et al., 2003). These bacteria are considered as generally recognized as safe (GRAS) microorganisms and can be safely used for medical and veterinary applications (Fuller, 1989). Lactobacilli are important organisms recognized for their fermentative ability as well as their health and nutritional benefits (Poorbaghi et al., 2014). They produce various compounds such as organic acids, diacetyl, hydrogen peroxide, bacteriocin and other bactericidal proteins during lactic fermentations (Ravi et al., 2011). Furthermore, Lactobacilli have been associated with numerous other health benefits such as the reduction of lactose intolerance (Fuller, 1991; Kiňová Sepová et al., 2008), increased immune response (Turroni et al., 2014; Villena et al., 2014), and anticancer properties (Viaud et al., 2014). In the food industry, LAB is widely used as starter cultures and has been cited to be part of human microbiota (Holzapfel et al., 2001). In raw milk and dairy products such as cheeses, yoghurts and fermented milks, Lactobacilli are naturally present or added intentionally, for technological reasons or to generate a functional food with health benefit for the consumers (Hassan and Frank, 2001).
Bacterial identification, including LAB, was performed by isolation and phenotypic study of the microorganism in the traditional microbiology laboratory. However, these methods have two major drawbacks: First, they require special equipment and expertise, second, some microorganisms exhibit biochemical characteristics that do not match the patterns for any known genus and species (Woo et al., 2002). Therefore, identification of Lactobacillus isolates by phenotypic methods is difficult, not reliable and some species are not readily distinguishable in terms of phenotypic characteristics (Afaf, 2012). According to above mentioned,derivation of simple and rapid identification methods is required in order to deal with the large numbers of Lactobacillus isolates obtained during microbial-ecological studies of ecosystems and also in food and health studies. Nucleotide base sequences of Lactobacillus 16S ribosomal DNA (16S rDNA) provide an accurate basis for phylogenetic analysis and identification (Amann et al., 1995; Nikolova et al., 2009). The sequence obtained from an isolate can be compared to those of Lactobacillus species held in data banks. On the other hand, the development of molecular techniques has opened up new perspectives for characterizing strains from fermented dairy foods (Tannock et al., 1999; Delfederico et al., 2006). Among PCR-based methods, 16S rDNA PCR- Restriction fragment length polymorphism (RFLP) analysis is easy, rapid and inexpensive way to identify microbial species such as yeast, acetic acid bacteria, and also few Gram positive bacteria (Claisse et al., 2007).
Iranian people in villages and nomad tribes make various fermented dairy products using cow and sheep milks. The transformation of surplus cow and sheep milk into traditional yoghurt is achieved through fermentation. The aims of the present study were to isolate and identify Lactobacillus existed in cow and sheep milk derived traditional yoghurt from the villages and nomad tribes of Fars province located in south of Iran. In this study, physiological, phenotypic and genotypic methods were used for accurate, valid and exact strain detection.
Materials and Methods
Isolation and culturing of microorganisms
Total 30 yogurt samples were collected randomly from nomad tribes of the different regions of Fars province, Iran and were used for the isolation of Lactobacillus strains. Sample collection was performed aseptically in sterile bottles kept in an ice-box, and transported immediately to the laboratory. One mL of each milk sample was homogenized with 9 mL of distilled water and mixed thoroughly for 60 S. Serial dilutions were made and aliquots (100 μL) of each dilution were streaked on MRS agar contained (g/L) 1% polypeptone, 1% meat extract, 0.5% yeast extract, 2% glucose, 0.5% sodium acetate, 0.2% ammonium citrate, 0.2% K2HPO4, 0.02% MgSO4.7H2O, 0.005% MnSO4.4H2O and 0.0108% Tween 80. It was incubated under anaerobic conditions for 48 hours at 37 °C.
Biochemical and probiotic characterization of the isolates
The morphological and physiobiochemical identification of bacteria were done according to Bergey’s Manual of Determinative Bacteriology (Holt, 1994). Colonies from highest dilution plates were randomly selected and purified by subculturing. Gram-positive, catalase negative and the most unique cultures were stored at -80 °C in MRS supplemented with 20% glycerol. Isolates were phenotypically assigned to the genus level on the basis of cell morphology, Gram staining, catalase-negative, indole negative and non-motility, according to the methods described previously (Gusils et al., 2004). Moreover, the acid production from carbohydrates was evaluated by using a miniaturized assay with glucose, D-galactose, D-mannitol, D-salicin,cellobiose, D-lactose, D-mannose, D-melibiose, maltose and D-lactose in microplates. After performing of all above mentioned tests, final and the most unique isolates were selected and genotypic methods of identification were carried out for them.
Molecular Identification of the Strains
DNA extraction and 16S rDNA gene sequencing were performed according to our previously published methods (Ghasemi et al., 2008; Rasoul-Amini et al., 2009). Briefly, 1mL of cultured cell was harvested by centrifugation (13000 g, 3 min at room temperature). Cells were resuspended in 0.5 mL of PBS buffer and the mixture was shaken slowly. The genomic DNA was extracted by heat shock method and used for PCR amplification of 16S rDNA gene. The 16S rDNA of the isolates was amplified using universal 16S ribosomal DNA primers: forward 5’-CAGCCGCGGTAATAC-3’ and reverse 5’-ACGGGCGGTGTGTAC-3’ which amplifiesa 800 bp region of the 16S rDNA gene. PCR reaction was performed in a total volume of 25 μL containing 200 μM dNTPs, 0.5 U Taq polymerase, 0.5 pM from each primers, 1 μL template, 50 mM MgCl2, 2.5 μL PCR buffer 10x and 19 μL DDW. Amplified DNA samples and a 100 bp ladder as molecular marker were electrophoresed in a 1% (w/v) agarose gel using tris borate EDTA (TBE) electrophoresis buffer containing 1μg/mL ethidium bromide. PCR products were purified and then sequenced by CinnaGen Company (Tehran, Iran).
Phylogenetics analysis of the amplified sequences
The resulting 16S rDNA gene sequences were aligned and compared to the nucleotide sequences of some known microorganisms in Gene Bank database of the National Center for Biotechnology Information (NCBI) by using Basic Local Alignment Tool (BLAST)(http://blast.ncbi.nlm.nih.gov/Blast.cgi).To create a multiple alignment, MAFFT multiple sequence alignment software version 7 (Katoh and Standley, 2013) was used. It uses a fast Fourier transform approach to align medium to large nucleotide sequences. The phylogenetic studies was conducted using MEGA software version 6 (Tamura et al., 2013). It was based on the 16S rDNA sequence of 800 bp drawn using the neighbor-joining method.Staphylococcus aurous was used as out-group strain.The CLC sequence viewer software (Qiagen, Aarhus, Denmark) version 7.5, was used to identify the conserved domains among these studied sequences.
Table 1: Biochemical and morphological tests on 20 selected Lactobacilli isolates
| Characteristics | Isolates | |||||||||||||||||||
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | |
| Carbohydrate fermentation | ||||||||||||||||||||
| Glucose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Cellobiose | + | – | + | – | + | + | + | + | – | + | + | + | + | + | + | + | + | – | + | + |
| Galactose | + | + | – | + | + | + | – | + | + | + | + | – | – | + | + | + | + | + | + | + |
| Lactose | + | – | + | – | – | + | – | + | + | + | – | + | + | – | + | + | + | + | + | + |
| Mannose | + | + | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | + | + |
| Mannitol | – | – | – | – | – | – | + | – | – | – | – | – | – | + | – | – | – | + | – | – |
| Maltose | + | + | – | + | + | + | + | + | + | + | – | + | + | + | + | + | + | + | + | + |
| Salicin | + | + | – | + | + | + | + | + | – | + | + | + | + | + | – | + | – | – | + | + |
| Melibiose | + | + | – | + | – | – | – | + | – | + | – | + | – | – | – | + | + | + | – | + |
| Sucrose | + | + | – | + | + | + | + | + | – | + | + | + | + | + | – | + | + | + | – | + |
| Catalase | – | – | – | – | – | + | – | – | – | ± | – | – | – | + | – | – | – | ± | – | – |
| Indole | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Motility | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
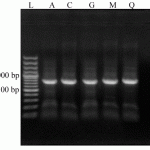 |
Figure 1: Agarose gel electrophoresis (1%) analysis of the amplified 16S rDNA sequences, extracted from five isolates (A, C, G, M and Q). Lane L: 100 bp DNA Ladder (Fermentas).A 800 bp 16S rDNA amplified band is distinguished for all isolates |
 |
Figure 2: The tabular format of a multiple alignment from five 16S rDNA genes of the isolatedLactobacillus strains and five related strains retrieved from the NCBI database, using the CLC sequence viewer software, version 7.5. |
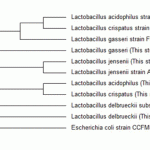 |
Figure 3: Molecular phylogenetic analysis of five isolated Lactobacillus strains with some related strains. |
Results
Collected yoghurt samples were combination of cow and sheep milk derived in approximately ratio of 2:1 (19 cow milk and 11 sheep milk derived yoghurt). From these 30 samples, Gram positive bacilli were detected only in 20 samples and among them 12 long, five mid to high and three short bacilli were existed. Results of biochemical and morphological test of 20 selected isolates are presented in Table 1. As demonstrated, all strains could utilize glucose, but 90% of them fermented maltose and mannose. Also, 80% of them fermented cellobiose, sucrose and galactose. The fermentative ability of these isolates was 75, 70, 50 and 15 percent for salicin, lactose, melibiose and mannitol, respectively. Results of catalase test of these isolates were 10%, 10% and 80% positive, negative and positive/negative, respectively. All isolates were indole negative and non-motile. From these isolates, totally five isolates were selected according to the most diversity of morphology and biochemical variables (isolates A, C, G, M, Q). All other molecular tests were performed only for these selected isolates. A band of 800 bp which represents 16S rDNA amplified sequence are shown in Figure 1. This band is specific for Lactobacillus genus and confirmed that these five selected isolates are belonged to Lactobacillus genus. Sequence analysis of this amplified bands and comparing them with NCBI database demonstrated a 98-99% of homology for each isolate with five distinct strains included L. acidophilus (isolate A, 98%), L. delbrueckii subsp. bulgaricus(isolate C, 99%), L. jensseni (isolate G, 98%), L. cripatus (isolate M, 98%) and L. gasseri (isolate Q, 98%).
The multiple sequence alignments, the similarity between the studied sequences and the conserved domains among the studied sequences were shown with a color scale (Figure2). The green residues are the least conserved and white residues are the most conserved.Sequence names appear at the beginning of each row and the residue position is indicated by the numbers at the top of the alignment columns. The consensus sequence is also shown in the below of ten studied sequences. Besides a yellow colored plot in percent scale, shows the conservation extent of each domain. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
The constructed phylogenetic tree is depicted in Figure 3. Molecular phylogenetic analysis of five isolated Lactobacillus strains with some related strains. The evolution history was inferred by using the Maximum Likelihood method. The bootstrap consensus tree inferred from 500 replicates. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The analysis involved 11 nucleotide sequences (accession numbers in parenthesis). All positions containing gaps and missing data were eliminated. Evolutionary analysis were conducted in MEGA6. Escherichia coli was used as out-group strain. It was revealed that there is a close genetic relationship between L. acidophilusandL. cripatus. This clade has a near relation with L. gasseri on its own. Whilst, L. delbrueckii showed just a long distant relationship with all other isolates.
Discussion
This study isolated and identified five strains of Lactobacillus from 30 different cow and sheep milk derived traditional yoghurt of villages and nomad population in Fars province, the biggest state of southern part of Iran. Our isolates were L. acidophilus, L. delbrueckii subsp. bulgaricus, L. jensseni, L. cripatus and L. gasseri. All of these strains potentially can be probiotic and help to health of the digestive tract and whole body of the host.
It must be remembered that conventional biochemical and physiological tests clearly have some limitations in discriminating between large numbers of isolates showing similar physiological characteristics (Zamfir et al., 2006; Mathara et al., 2008). The strategy described in this study is an effective means of identifying unknown strains of the Lactobacillus complex, which is a critical initial step in the selection and development of novel probiotic strains. Similar strategy was used in different previous studies worldwide (Kullen et al., 2000; Lee et al., 2008; Macías-Rodríguez et al., 2008). Also, in Iran, there are several reports about using this methods for identifying and selecting appropriate probiotic strains. In a study conducted by Latifi et al, LAB were isolated and characterized using phenotypic methods (Gram staining, physiological and biochemical tests) from traditional cheese and yoghourt of Heris and Sarab regions. Then their acid and bile tolerance, as the primary probiotic characterizations, were investigated. 16S rDNA gene of Lactobacilli and Enterococci was amplified for identification of bacterial strains. Totally, 15 Lactobacillus spp. and 16 Enterococcus spp. were isolated from traditional dairy products of these regions that could be potentially used in the industrial dairy products (Latifi et al., 2010). In anotherpublished study, 17 strains of Lactobacillus were isolated from different Iranian dairy products. Their study showed the highest genetic diversity in yoghurt population. Also, all strains had 99% genetic similarity with L. casei(Tafvizi and Tajabadi Ebrahimi, 2012). Isolation and identification of Lactobacilli in traditional fermented milk from two different provinces in the west of Iran were carried out by Dana et al. Lactobacillus bacteria were isolated, and characterized phenotypically. The 16S rDNA genes from these two strains were amplified and sequenced and a phylogenetic tree was constructed. The sequencing results in combination with phenotypic and biochemical properties showed that both strains were similar to L. crustorum(Dana et al., 2013).
Some differences were seen between biochemical tests (especially carbohydrate fermentation tests) in this study and registered properties for detected strains. These differences may be due to some mutations in the genes that responsible for metabolic enzymes. Further studies are needed for finding these interfering mutations and evaluation of their relationship with probiotic beneficial properties.
Conclusion
According to our findings, phylogenetic analysis using 16S rDNA sequences appeared to be a very practical method and highly sensitive in the discrimination of the Lactobacillus species. Also, this study demonstrated that LAB (especially Lactobacillus genus) are the most frequently probiotics which are found in traditional Persian yoghurt.
Acknowledgements
The authors would like to thank Research Deputy of Shiraz University of Medical Sciences, Shiraz, Iran for the financial support of this project. This study was a part of Pharm. D. thesis of Mohammad Hadi Zare, proposed and approved in Faculty of Pharmacy, Shiraz University of Medical Sciences, Shiraz, Iran.
References
- Afaf, I. S.Molecular identification of probiotics Lactobacillus strain isolates by amplified ribosomal DNA restriction analysis (ARDRA). J.Microbiol. Res.,2012;6:3034-3041.
- Amann, R.I., Ludwig, W., Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Rev.,1995;59:143-169.
- Claisse, O., Renouf, V., Lonvaud-Funel, A. Differentiation of wine lactic acid bacteria species based on RFLP analysis of a partial sequence of rpoB J. Microbiol. Methods,2007;69:387-390.
- Coeuret, V., Dubernet, S., Bernardeau, M., Gueguen, M., Vernoux, J.P. Isolation, characterisation and identification of lactobacilli focusing mainly on cheeses and other dairy products. Lait, 2003; 83:269-306.
- Dana,M.G., HatefSalmanian, A., Yakhchali, B., RastegarJazi, F. High folate production by naturally occurring Lactobacillus with probiotics potential isolated from dairy products in Ilam and Lorestan provinces of Iran. Afr. J. Biotechnol.,2013;9:5383-5391.
- Delfederico,L., Hollmann, A., Martínez, M., Iglesias, N.G., De Antoni, G., Semorile, L. Molecular identification and typing of lactobacilli isolated from kefir grains. Dairy Res., 2006;73:20-27.
- Fuller, R. Probiotics in man and animals. Appl. Bacteriol., 1989;66:365-378.
- Fuller, R. Probiotics in human medicine. Gut,1991; 32:439-442.
- Ghasemi, Y., Rasoul-Amini,S., Morowvat, M.H., Raee, M.J., Ghoshoon, M. B., Nouri, F., Negintaji, N., Parvizi, R., Mosavi-Azam, S. B. Characterization of hydrocortisone biometabolites and 18S rRNA gene in Chlamydomonas reinhardtii Molecules, 2008;13:2416-2425.
- Gusils,C., Chaia, A. P., Oliver, G., González, S. Microtechnique for identification of lactic acid bacteria. Methods Mol. Biol., 2004; 268:453-458.
- Hassan, A. N., Frank, J. F. Starter cultures and their use. Food Sci.Tech.,2001; 151-206.
- Holt, J. G.(ed): Bergey’s Manual of Determinative Bacteriology,9th Philadelphia:LWW, 1994.
- Holzapfel, W. H., Haberer, P., Geisen, R., Björkroth, J., Schillinger, U. Taxonomy and important features of probiotic microorganisms in food and nutrition. J. Clin. Nutr.,2001;73:365S-373S.
- Jothi, V. V., Anandapandian, K., Shankar, T. Bacteriocin production by probiotic bacteria from curd and its field application to poultry. Appl. Sci. Res.,2012; 4:336-347.
- Katoh, K., Standley, D. M.MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Biol. Evol.,2013; 30:772-780.
- Kiňová Sepová, H., Bilková, A., Bukovský,M. Lactobacilli and their probiotic properties. Ceska Slov. Farm., 2008; 57:95-98.
- Kullen, M. J., Sanozky-Dawes, R. B., Crowell, D. C., Klaenhammer, T. R.Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus J. Appl.Microbiol.,2000; 89:511-516.
- Latifi, H., Hejazi, M., Maleki Zanjani, B., Barzegari, A. Isolation, biochemical and molecular identification of potentially probiotic bacteria from traditional dairy products from Heris and Sarab regions. Food Res., 2010; 3:1-17.
- Lee, C., Sieo, C., Wong, C., Abdullah, N., Ho, Y. Sequence analysis of 16S rRNA gene and 16S–23S rRNA gene intergenic spacer region for differentiation of probiotics Lactobacillus strains isolated from the gastrointestinal tract of chicken. Microbiol.,2008; 58:133-140.
- Macías-Rodríguez,M., Zagorec, M., Ascencio,F., Rojas, M. Potential probioticLactobacillus strains for piglets from an arid coast. Microbiol., 2008; 58:641-648.
- Mathara, J.M., Schillinger, U., Guigas, C., Franz, C., Kutima, P. M., Mbugua, S. K., Shin, H. K., Holzapfel, W. H. Functional characteristics of Lactobacillus from traditional Maasai fermented milk products in Kenya. Int. J. Food Microbiol., 2008;126:57-64.
- Nikolova, D., Evstatieva, Y., Georgieva, R., Danova, S., Savov, V., Ilieva, S., Dalev, P. Molecular taxonomic characterisation of probiotic strain Lactobacillus 50P1. Biotechnol.Biotechnol. Equip.,2009; 23:779-782.
- Poorbaghi, S. L., Dadras, H., Gheisari, H. R., Mosleh, N., Firouzi, S., Roohallazadeh, H. Effects of Lactobacillus acidophilus and inulin on faecal viral shedding and immunization against H9N2 Avian influenza virus. Appl. Microbiol.,2014;116:667-676.
- Rasoul-Amini, S., Ghasemi, Y., Morowvat, M. H., Mohagheghzadeh, A. PCR amplification of 18S rRNA, single cell protein production and fatty acid evaluation of some naturally isolated microalgae. Food Chem., 2009; 116:129-136.
- Ravi, V., Prabhu, M., Subramanyam, D. Isolation of bacteriocin producing bacteria from mango pulp and its antimicrobial activity. Microbiol. Biotech. Res.,2011; 1:54-63.
- Tafvizi, F., Tajabadi Ebrahimi, M. Detection of genetic diversity and classification of Lactobacillus species isolated from Iranian traditional dairy products by RAPD fingerprinting and POPGENE analysis. Biol. Res.,2012; 3:4904-4911.
- Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Biol.Evol.,2013; 30:2725-2729.
- Tannock, G. W., Munro, K., Harmsen, H. J. M., Welling, G. W., Smart, J., Gopal, P. K. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus Appl. Env. Microbiol., 2000; 66:2578-2588.
- Tannock, G. W., Tilsala-Timisjarvi, A., Rodtong, S., Ng, J., Munro, K., Alatossava, T. Identification of Lactobacillus isolates from the gastrointestinal tract, silage, and yoghurt by 16S-23S rRNA gene intergenic spacer region sequence comparisons. Env. Microbiol.,1999; 65:4264-4267.
- Turroni, F., Taverniti, V., Ruas-Madiedo, P, Duranti, S., Guglielmetti, S., Lugli, G. A., Gioiosa, L., Palanza, P., Margolles, A., Van Sinderen, D., Ventura, M.Bifidobacterium bifidum PRL2010 modulates the host innate immune response. Env. Microbiol.,2014; 80:730-740.
- Viaud, S., Daillère, R., Boneca, I. G., Lepage, P., Langella, P., Chamaillard, M., Pittet, M. J., Ghiringhelli, F., Trinchieri, G., Goldszmid, R., Zitvogel, L. Gut microbiome and anticancer immune response: really hot Sh*t! Cell Death Differ.,2014; 1-16.
- Villena, J., Chiba, E., Vizoso-Pinto, M. G., Tomosada, Y., Takahashi, T., Ishizuka, T., Aso, H., Salva, S., Alvarez, S., Kitazawa, H. Immunobiotic Lactobacillus rhamnosus strains differentially modulate antiviral immune response in porcine intestinal epithelial and antigen presenting cells. BMC Microbiol., 2014; 14.
- Woo, P. C. Y., Fung, A. M. Y., Lau, S. K. P., Yuen, K. Y. Identification by 16S rRNA gene sequencing of Lactobacillus salivarius bacteremic cholecystitis. Clin. Microbiol., 2002; 40:265-267.
- Zamfir, M., Vancanneyt, M., Makras, L., Vaningelgem, F., Lefebvre,K., Pot, B., Swings, J., De Vuyst, L. Biodiversity of lactic acid bacteria in Romanian dairy products. Syst. Appl. Microbiol.,2006; 29:487-495

This work is licensed under a Creative Commons Attribution 4.0 International License.





