How to Cite | Publication History | PlumX Article Matrix
Differences in protease expression of mutants Bacillus licheniformis F11
Ace Baehaki1*, Dahrul Syah2, Sukarno2, Siswa Setyahadi3, and Maggy T. Suhartono3
1Department of Fisheries Product Technology, Faculty of Agriculture, Sriwijaya University, Indralaya, South Sumatera, Indonesia 2Department of Food Science and Technology, Bogor Agricultural University, Darmaga Campus IPB Bogor 16002, Indonesia 3Agency for the Assessment and Application of Technology, Jakarta, Indonesia
DOI : http://dx.doi.org/10.13005/bbra/1871
ABSTRACT: The objective of the study was to investigate the differences in protease productions of mutans Bacillus licheniformis F11. In this study B. licheniformis F11.1 (DchiA; Dpga) and B. licheniformis F11.4 (DchiAB; Dpga) was used. Two types (LB and MLB + collagen) of media were used for bacterial growth, protease production and zymogram analysis : Luria Broth (LB) and Modified Luria Broth plus collagen (MLB +collagen) media. The LB media contained tryptone 1%, NaCl 1% dan yeast extract 0.5%. The Modified LB + collagen media contained tryptone 0.5%; NaCl 1%, yeast extract 0.25% and collagen 5%. Two protease producing B. licheniformis F11 mutans were screened from Palembang Indonesia, Bacillus licheniformis F11.1 (DchiA; Dpga) and Bacillus licheniformis F11.4 (DchiAB; Dpga). Both mutants showed different growth pattern and protease production in media containing collagen. F11.1 displays a higher protease activity than F11.4 in Luria Bertani media with an optimum after 15 hours for F11.1 and 15-25 hours for F11.4. Upon addition of collagen to LB medium F11.4 overmatched F11.1 with respect to collagenolytic activity. The presence of collagen led to a shift in the substrate preference of the two strains towards collagen compared to other protein substrates (gelatin, fibrin and casein). Zymogram analysis on gel embedded substrates suggests a higher molecular variation for strain F11.4.The different genetic mutation of the same species (Bacillus licheniformis F11.1 and F11.4) displayed different protease production patterns and substrate specificities especially on collagen.
KEYWORDS: Protease; Bacillus licheniformis F11.1; F11.4; Bacillus licheniformis F11.1; F11.4; expresion
Download this article as:| Copy the following to cite this article: Baehaki A, Syah D, Sukarno, Setyahadi S, Suhartono M. T. Differences in protease expression of mutants Bacillus licheniformis F11. Biosci Biotechnol Res Asia 2015;12(3) |
| Copy the following to cite this URL: Baehaki A, Syah D, Sukarno, Setyahadi S, Suhartono M. T. Differences in protease expression of mutants Bacillus licheniformis F11. Biosci Biotechnol Res Asia 2015;12(3). Available from: https://www.biotech-asia.org/?p=3517> |
Proteases are involved in numerous cellular operations, such as protein metabolism, post translational modifications, extracellular enzyme secretion, sporulation, and even blood coagulation1,2. There are non specific proteases, which find applications as washing-detergents as well as in the leather and food industry and for waste processing. Specific proteases are applied as medical agents for treating diseases such as indigestion, artherosclerosis, stroke, as cosmetic-ingredients but also for molecular biological purposes. Application of specific proteases to marine and plant proteins were shown to be important for the production of a panoply of bioactive molecules such as peptides with antiviral, antioxidant and inhibitory effects of the Angiotensin Converting Enzyme3-5.
Microbial proteolytic enzymes are classified as serine proteases (E.C.3.4.21), sulfhydril proteases (E.C.3.4.22), metallo proteases (E.C.3.4.24) and acid proteases (E.C.3.4.23). Though several microorganism are known as potent protease producers : Bacillus subtilis CN26, Bacillus sp.7, and Bacillus subtilis PE-118. Representatives of the genus Bacillus constitute the major bacterial workhorses for commercial enzyme production. Bacilli encode a great number of secreted enzymes, including proteases, amylases, glucanases, etc9. Genes encoding secreted enzymes have been cloned and functional analyses as well as genetic regulation was studied in detail as for Bacillus megaterium10-12. The ubiqitiously occurring Bacillus licheniformis (in many respects a Generally Regarded As Safe or GRAS organism) plays the most prominent role since it produces up to 25 g/l extracellular proteins6. The availability of its genome sequence further facilitates use of B. licheniformis as a valuable enzyme producer13,14.
In a search for protease producing microorganisms which can be applied for deproteination of chitin samples derived from shrimp waste, Bacillus licheniformis F11 was isolated from shrimp waste in Palembang and characterized as the most promising candidate15. Amino acid alignments of predicted chitinases revealed 99 to 100% identity to the known B. licheniformis enzymes , however, a frameshift mutation was detected in chiA of the strain favouring the strain for the above purpose as it almost completely lacks chitinase activity but hypersecretes proteases15. By a targeted gene deletion approach two mutants of B. licheniformis F11 were generated: (1) B. licheniformis F11.1 which in addition to the above frame shift mutation in chiA lacks the genes for polyglutamte synthesis (Dpga), and (2) F11.4 which has the Dpga deletion and is completely lacking both of the chitinase encoding genes DchiBA16.
Here, we report findings that the twoalmost isogenic B. licheniformis strains F11.1 and B. licheniformis F11.4 display different protease production patterns and substrate specificities especially on collagen.
Material and Methods
Bacterial Strains and Growth Conditions
In this study we used B. licheniformis F11.1 (DchiA; Dpga) and B. licheniformis F11.4 (DchiAB; Dpga). For detailed description we refer to Waldeck et al.15. Two types of media were used for bacterial growth and protease production : Luria Broth (LB) and Modified Luria Broth plus collagen (MLB +collagen) media. The LB media contained tryptone 1%, NaCl 1% and yeast extract 0.5%. The Modified LB + collagen media contained tryptone 0.5%; NaCl 1%, yeast extract 0.25% and collagen 5%. Cell growth was monitored turbidimetrically by measuring the absorbance at λ = 620 nm. 10 % of the seedling culture with an OD620 of 0.8 was inoculated into the same media for enzyme production. Incubation was performed at 37 0C and samples were taken at regular intervals for analysis of enzyme activity and cell growth.
Protein Assay and Protease Activity
The protein concentration was measured according to Bradford17 using reagents consisting of 100 mg Coomassie brilliant blue (CBB) G-250 in 50 ml 95 % ethanol and 100 ml 85 % phosphate acid in 1 liter. Bovine serum albumin served as the protein standard. Each experiment was done in triplicate.
Protease activity was measured according to the method proposed by Bergmeyer et al.18 using either casein, collagen, gelatin or fibrin at 1 % w/v concentration in buffer Tris-HCl 0.05 M. 50 μl enzyme filtrate was mixed with 250 μl substrate and incubated for 10 minutes at 37 0C. Trichloracetic acid (TCA) 0.2 M was added and kept at 37 0C for 10 minutes followed by centrifugation (4000xg,10 minutes). The supernatant was mixed with 0.4 M Na2CO3, followed by addition of Folin Ciocalteau (reagent (1:2) and further incubation at 37 0C for 20 minutes. The reaction products were measured at λ 578 nm. Substrate solution without enzyme was used as control. One unit (U) of enzyme activity equals 1 μ mole tyrosine per min.
Zymogram Analysis
For zymogram analysis, a modified SDS-PAGE procedure was applied. The polyacrylamide gel used were 8 % for separating gel and 4% for collecting (modified after Lantz and Ciborrowski19. Acrylamide was copolymerized with 1% of the protein substrates. Following electrophoresis, the gel was soaked in Triton X-100 2.5% for 1 hour and further incubated for enzyme substrate reaction in Tris-Cl 10 mM, pH 8 for 24 hours. The activity bands were visualized after (50 % methanol + 10 % acetic acid + 0,06 % Coomassie brilliant blue R-250) for 30 minutes followed by incubation in destaining solution (5 % methanol + 7,5 % acetic acid). Positive results corresponded to clear bands. Estimation of the molecular weight was conducted by using Low Molecular Weight protein standard phosphorylase b (97 kDa), BSA (bovine serum albumin) (66,0 kDa), ovalbumin (45 kDa), carbonic anhydrase (30 kDa), and lysozyme (14.3 kDa).
Results
The Bacillus lichneiformis F11 showed high protease activity but lacking chitinase activities. Morphology characteristics was conducted previously and the F11 displayed a rough colony morphology, and motile with rods shape. Further physiological and biochemical characterization suggested representatives of the Bacillus licheniformis species. Definite evidence for the affiliation of the isolate with the species B. licheniformis was obtained from sequencing the 16S rRNA gene which indicated 100% identity to B. Licheniformis DSM13/ATCC 14580. The genetic analysis had included a number of loci encoding extracellular degradative enzymes, including adjacent regions, and the degS–degU operon instrumental in the regulation of such enzymes, as well as the pga locus, which encodes the polyglutamic acid synthetase. When a Blastp search was performed with the deduced polypeptides, 99 to 100% identity to known Bacillus licheniformis loci became evident, with only one exception, i.e., chiA15. By aligning the corresponding predicted proteins and DNA sequences it became evident that there is a frameshift mutation in chiA in F11.1 and there is another additional mutation in chiB in F11.416. In this research, different enzyme expression and pattern from both isolates were obtained.
The strains B. licheniformis F11.1 and F11.4 differ genetically only with respect to their chitinase genes, the former has the frame shift mutation in chiA while the latter has the chitinase encoding genes chiBA completely deleted. The Dpga deletion is common to both mutants, it was introduced to save stirrer energy during fermentation (reduced viscosity) and no metabolic energy is wasted for an unwanted by-product. Since both strains performed rather differently in deproteinization experiments with shrimp shells15,16. We were eager to learn more about their capacity to produce proteolytic enzymes.
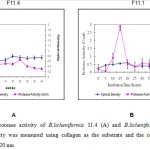 |
Figure 1: Protease activity of B.licheniformis 11.4 (A) and B.licheniformis 11.1 (B) |
As seen as Figure 1 showed that B.licheniformis 11.1 produced higher protease activity compared to the B. licheniformis F11.4. The optimum production time was 15 for B. lichneiformis F11.1 with a protease activity of 0.028 U/ml. For B. licheniformis F11.4 it was between 20 – 35 hours of fermentation, with enzyme activity of 0.005 U/ml. After 15 hours in LB media, the protease activity of B. licheniformis F11.1 decreased significanty. The decreasing enzyme activity might be due to autohydrolysis of the protease molecules. The lower protease activity of B. licheniformis F11.4 appeared to correlate with the more stable and longer period of appearance of enzyme activities.
Earlier study by Hoffmann et al.16 who used Luria Bertani media with azocasein as the substrate reported similar result, that is, B. licheniformis F11.1 showed shorter time for protease synthesis (20 hours), and B. licheniformis F11.4 required longer time of 40 hours to optimally produced its protease. In LB media, the protease activity of B. licheniformis F11.1 was higher than that of extracellular B. licheniformis F11.4 . The extracellular protease from B. licheniformis F11.1 was effective for deproteinization of the protein attached to the shrimp waste. The collagenase activity of Bacillus licheniformis F11.4 was reported20. Addition of collagen into the media LB, resulted in different responses. In this case, the concentration of tryptone and yeast extract in the original LB media was reduced by half, and collagen was added at 5%., the activity of Bacillus licheniformis F11.4 was increased20.
 |
Figure 2: Activity of protease from Bacillus licheniformis 11.4 (A and C) and Bacillus licheniformis 11.1 (B and D) produced in media LB (A and B) and media modified LB + Collagen (C and D). |
Figure 2 showed difference pattern substrate preferences of protease from B.licheniformis F11.1 and B. licheniformis F11.4. When produced in LB media, both protease were more active towards casein compare with the other 3 protein substrates (collagen, gelatin and fibrin). Furthurmore, when gelatin was used as the substrate for enzyme analysis, the protease from B.licheniformis F11.4 showed higher enzyme activity up to 3 times higher that than of B.licheniformis F11.1. Addition of collagen into the media induced higher synthesis of fraction capable of degrading collagen. Zymography analysis in collagen gel (Figure 3) showed increase not only in the number of clearing band but also more intense clearing band.
 |
Figure 3: Activities of protease fractions from B. licehniformis F11.4 (A and C) and B |
Figure 3 and Table 1 confirmed the different protease fractions produced by both B. licehniformis F11 mutants grown in different media. Diversity of the protease fractions with different apparent molecular weight were detected through zymography analysis. B. licheniformis F 11.4 synthesized protease fraction with higher molecular weight on the four substrates tested (casein, collagen, gelatin and fibrin). Table 1 also indicated that the protease fraction produced by B.licheniformis F11.4 in the collagen containing media were more active toward not only collagen but to other protein substrates as well.
Table 1: Summary of protease fractions produced by B. licehniformis F11.4 and B. licehniformis F11.1 detected in zymogram analysis
| MW | F11.4 | F11.1 | ||||||
| Casein | Collagen | Gelatin | Fibrin | Casein | Collagen | Gelatin | Fibrin | |
| < 40 | √ | √ | √ | |||||
| 40-90 | √ | √ | √ | √ | ||||
| >90 | √ | √ | √ | √ | √ | √ | √ | |
Discusion
The optimum production time was 15 hours for B. licheniformis F11.1 and 12-35 hours for B. licheniformis F11.4. The higher enzyme activity might be responsible to the enzyme autohydrolysis which is shown as quick reduction in the enzyme activity following it is optimum level. This is different from the protease from B. licheniformis F11.4 which remained high in the LB media at longer time intervals. For B. licheniformis F11.1, the optimum enzyme was produced during logarithmic phase and also reached maximum activity at the time close to stationary phase.
Generally, extracellular proteases are produced by Bacillus sp. during post exponential and stationary phases. Little or no enzyme production had been reported during exponential growth phase of Bacillus; however protease production in B. subtilis ATCC14416 and B. sphaericus BSE 18 occurs at mid-exponential phase, and rapid auto deactivation was observed after the culture reached maximum enzyme activity. There is case which reported that exponential growth phase was the phase in which the synthesis and secretion of the protease was initiated, with a substantial increase near the end of the growth phase and maximum amounts of protease produced in the stationary growth phase21.
The optimum fermentation time for synthesis of a particular enzyme is affected not only by different bacterial strain but also by varieties of fermentation conditions. In general, the Bacillus subtilis CN2 was reported to synthesize protease optimally by 14 hours of fermentation time6. Shorter optimum fermentation time of protease, ie. 9 hours was reported when Bacillus SMIA-2 was grown in media containing trisodium citrate 1%7. Bacillus subtilis PE-11 produced protease optimally at longer incubation time, that by 48 hours when grown in nutrient broth8.
In collagen media, the extracellular protease from both B. licheniformis were more active toward collagen eventough casein was still best substrate. Peptide bond in casein structure are more accessible for enzyme hydrolysis, while more complex three dimensional fibril structure of other substrates contribute to reduced chance of enzyme substrate interaction. Due to their triple helical nature, collagen and gelatin is usually resistant to many general proteases21. Protease from Bacillus also showed preferences to protein substrate like casein, albumin and less active to the more complex protein such as collagen, keratine and fibrin22.
Excretion of several protease molecules which are capable of degrading different substrates is a general phenomenon found in many bacteria and microorganisms. This is essential to utilize the numerous protein substrates found in nature1,2. Nowadays, multi proteases which are able to degrade different protein substrates such as gluten and casein is much applied as agents to help indigestion problems. In our study, beside capable of digesting casein and collagen, protease of Bacillus licheniformis F 11.1 and F 11.4 could hydrolyse gelatin and fibrin. Another protein which was found as substrate for the enzymes was wheat derrived gluten known as protein which cause alergenicity (data not shown). Protease treatment is one of the many proposed techniques useful to reduce some allergenic responses.
Conclusions
The presence of collagen in the production media clearly induced enzyme fractions which were more active towards collagen. Zimography assay is thus useful to get more information on the enzyme molecular fractions synthesized at various conditions. The different genetic mutation of the same species (Bacillus licheniformis F11.1 and F11.4) results in different metabolic activities which result in difference characteristic of the protease molecules synthesized.
Acknowledgements
This research was funded by Indonesian Ministry of Education and Culture through 2010 International Research Scheme. Bacillus licheniformis F11 used in this research were the result of research collaboration between Indonesia (Agency for the Assessment and Application of Technology Jakarta ) and German (Indo-German Biotechnology). We gratefully acknowledge for Prof. Dr. Friedhelm Meinhardt and his colleague for constructed of the mutant Bacillus licheniformis F11.1 and F11.4
References
- Rao, M.B., Tanksale, A.M., Ghatge, M.S., Deshpande V.V. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol., 1998; 62:597-635.
- Ward, O.P., Rao, M.B., Kulkarni, A. Proteases production. In: Encylopedia of Microbiology (Schaechter M, ed). United States of America: Elsevier, 2009; pp. 495-511,
- Sasaki, T., Larrson, H., Tisi, D., Claesson-Welsh, L., Hohenester, E., Timpl, R. Endostatin derived from collagens XV and XVIII differ in structural and binding properties, tissu distribution and antiangiogenic activity. J. Mol. Biol., 2000; 301: 1179-1190.
- Klompong, V., Benjakul, S., Kantachote, D., Shahidi, F. Antioxidative accctivity and functional properties of protein hydrolysate of yellow stripe trevally (Selaroides leptolepis) as influenced by the degree of hydrolysis and enzyme type. Food Chem., 2007; 102: 1317-1327.
- Liu, B.L., Chiang, P.S. Production of hydrolysate with Antioxidative actiovity and functional properties by enzymatic hydrolysis of defatted sesame (sesame indicum L.). Int. J. Appl. Scie. Eng., 2008; 6: 73-83.
- Tran, I.H., Nagano, H. Isolation and characteristic of Bacillus subtilis CN2 and its collagenase production. J. Food Sci., 2002; 67: 1184-1187.
- Nascimento, W.C.A., Martins, M.L.L. Production and properties of an extracellular protease from thermophilic Bacillus Braz. J. Microbiol., 2004; 35: 91-96.
- Adinarayana, K., Sllaiah, P., Prasad, D.S. Production and partial characterization of thermostable serine alkaline protease from a newly isolated Bacillus subtilis PE-11. AAPS. PharmSciTech., 2003; 4: E56-64.
- Schallmey, M., Singh, A., Ward, O.P. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol., 2004; 50: 1-17.
- Meinhardt, F., Bußkamp, M., Wittchen, K.D. Cloning and sequencing of the leu C and npr M genes and a putative spo IV gene from Bacillus megaterium DSM 319. Appl. Microbiol. Biotechnol., 1994; 41: 344-351.
- Strey, J., Wittchen, KD., Meinhardt, F. Regulation of b-galactosidase expression in Bacillus megaterium DSM 319 by a XylS/AraC-type transcriptional activator. J. Bacteriol., 1999; 181: 3288-3292.
- Lee JS, Wittchen KD, Stahl C, Strey J, Meinhardt. F. Cloning, expression and carbon catabolite repression of the bamM gene encoding b-amylase of Bacillus megaterium DSM 319. Appl. Microbiol. Biotechnol., 2001; 56: 205-211.
- Veith, B., Herzberg, C., Steckel, S., Feesche, J., Maurer, K.H., Ehrenreich, P., Baumer, S., Henne, A., Liesegang, H., Merkl, R., Ehrenreich, A., Gottschalk, G. The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. Mol. Biotechnol., 2004; 7: 204-211.
- Rey, M.W., Ramaiya, P., Nelson, B.A., Brody-Karpin, S.D., Zaretsky, EJ., Tang, M., Lopez de-Leon, A., Xiang, H., Gusti, V., Clausen, I.G., Olsen, P.B., Rasmussen, M.D., Andersen, J.T., Jorgensen, P.L., Larsen, T.S., Sorokin, A., Bolotin, A., Lapidus, A., Galleron, N., Ehrlich, S.D., Berka, R.M. Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome. Biol., 2004; 5: R77.
- Waldeck, J., Daum, G., Bisping, B., Meinhardt, F. Isolation and Molecular Characterization of Chitinase-Deficient Bacillus licheniformis Strains Capable of Deproteinization of Shrimp Shell Waste To Obtain Highly Viscous Chitin. Appl. Environ, Microbiol., 2006; 72: 7879–7885.
- Hoffmann, K., Daum, G., Koster, M., Kulicke, W.M., Meyer-Rammes, H., Bisping, B., Meinhardt, F. Genetic improvement of Bacillus licheniformis strains for efficient deproteinization of shrimp shells and production of high-molecular-mass chitin and chitosan. Appl. Environ. Microbiol., 2010; 76: 8211-8221.
- Bradford, M.M. A rapid and sensitive methode for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem., 1976; 72: 248-254
- Bergmeyer, H.U., Grabl, M., Walter, H.E. Enzymes. In: Methods of Enzymatic Analysis (Bergmeyer HU, Grapl M, eds.). Weinheim: Verlag Chemie, 1983; pp.1007-1009,
- Lantz, M.S., Ciborowski, P. Zymographic techniques for detection and characterization of microbial proteases. Methods Enzymol., 1983; 235: 563-594.
- Baehaki, A., Sukarno., Syah, D., Setyahadi, S., Suhartono, M.T. Production and Characterization of Collagenolytic Protease from Bacillus licheniformis F11.4 Originated from Indonesia. Asian J. Biochem., 2014; 26 (10): 2861-2864.
- Kumar, C.G, Hiroshi T. Microbial alkaline proteases: From a bioindustrial viewpoint. Biotechnol. Adv., 1999; 17: 561-594.
- Chung, L., Dinakarpandian, D., Yoshida, N., Lauer-Fields, J., Fields, G.B., Visse, R., Nagase, H. Collagenase unwinds triple helical collagen proir to peptide. EMBO J., 2004; 23: 3020-3030.

This work is licensed under a Creative Commons Attribution 4.0 International License.





