How to Cite | Publication History | PlumX Article Matrix
Priya Pradhan*, Nishant K. Soni, Lalitesh Chaudhary, Somdutt Mujwar and K. R. Pardasani
Department of Bioinformatics, Maulana Azad National Institute of Technology Bhopal 462 051 (INDIA)
DOI : http://dx.doi.org/10.13005/bbra/1889
ABSTRACT: Almost every age group is at higher risk for serious flu complications. The major problem arising these days regarding the control of influenza disease is the development of resistance among the influenza viruses against the existing anti-viral drugs that are being recommended. Also, these antiviral drugs have a number of side effects. The main objective of the present paper is to explore riboswitches as a novel target for design of drugs for influenza virus in order to address the issues of resistance and side effects of present drugs. Riboswitches are present in the non-coding region of mRNA that sense changes in the cellular environment and directly mediate appropriate gene control responses. These riboswitches are primarily found in the 5’ untranslated regions of messenger RNAs. In the present paper in-silico approach is proposed for the prediction of riboswitches for the strains of influenza virus, their binding sites and design of their inhibitors. Two riboswitches have been predicted for the three strains of influenza virus and five inhibitors have been identified for each of the two riboswitches by virtual screening. These inhibitors are found to be free from the side effects of antiviral agents and have remote chances of being resistant.
KEYWORDS: Riboswitches; Influenza; Inhibitors; Auto dock; Drug; Virtual Screening
Download this article as:| Copy the following to cite this article: Pradhan P, Soni N. K, Chaudhary L, Mujwar S, Pardasani K. R. In-Silico Prediction of Riboswitches and Design of their Potent Inhibitors for H1N1, H2N2 and H3N2 Strains of Influenza Virus. Biosci Biotechnol Res Asia 2015;12(3) |
| Copy the following to cite this URL: Pradhan P, Soni N. K, Chaudhary L, Mujwar S, Pardasani K. R. In-Silico Prediction of Riboswitches and Design of their Potent Inhibitors for H1N1, H2N2 and H3N2 Strains of Influenza Virus. Biosci Biotechnol Res Asia 2015;12(3). Available from: https://www.biotech-asia.org/?p=2454 |
Introduction
Influenza is a contagious disease affecting the respiratory tract, caused by Influenza viruses A, B, C mainly A, its symptoms can lead from mild to severe illness and at times can lead to death as compared to typical sneezing and stuffiness of common cold. The flu viruses spread mainly by droplets when people with the flu cough, sneeze or talk. These droplets mainly land in the mouths or noses of people who are in the vicinity [1-7]. Moreover, the flu may get spread over an area directly or indirectly.
Symptoms of Influenza are Fever, Cough, Sore throat, Runny nose, Muscle or body aches, Headaches, Fatigue and some people may have vomiting and diarrhea also [8,9]. Centers for Disease Control and Prevention estimated that around 90% of seasonal flu related deaths and more than 60% of hospitalizations in the united states occur in people of 65 years of age and older because human immune system becomes weaker with age [10].
People suffering with asthma have swollen and sensitive airways, and when they experience symptoms of influenza, it can cause further inflammation of the airways and lungs, not only asthmatic patients but also people with neurological disorders [11] suffer badly with influenza. People suffering with chronic lung diseases, Blood disorders (such as sickle cell disease), Endocrine disorders (such as diabetes mellitus), Kidney disorders, Liver disorders, people with HIV or AIDS and heart disease are also at high risk for flu complications.
The major problem with the existing antiviral drugs is the development of resistance among the influenza viruses against these drugs. Earlier Adamantanes i.e. Amantadine and Rimantadine were given for influenza treatment, but it was observed that resistance among influenza a viruses increased so fast for Adamantanes that it became necessary to stop their use in the treatment. It was observed that Adamantane resistance increased from 0.4% during 1994-1995 to 12.3% during 2003-2004 [12].
After this treatment Neuraminidase Inhibitors i.e. Oseltamivir or zanamivir that are the primary antiviral agents were recommended for the prevention and treatment of influenza viruses [13-15]. But later the development of resistance [16-19] for these antiviral drugs was reported which states that oseltamivir-resistant seasonal influenza A viruses were isolated from nine (18%) out of 50 Japanese children during treatment [17,20]. Zanamivir is not recommended for immune-compromised children [16,21]. During the post marketing surveillance allergic reactions, facial edema, and swelling are also reported [22,23]. Apart from the development of resistance, the antiviral drugs have number of side effects.
The major problem faced with vaccination i.e. flu shots is that it might have some of the common local adverse reactions that includes erythema (redness), induration (firmness), swelling, pain, and pruritus (itching) at the vaccination site; headache, myalgia (muscle ache), and malaise.
These issues of drugs resistance and side effects of existing drugs are of major concern for treatment of influenza virus and there thus there is a need to explore novel drugs targets and design new inhibitor for influenza virus. The riboswitches can be explored as new drug target for such infectious diseases.
Riboswitches are present in the non-coding region of mRNA that sense changes in the cellular environment and directly mediate appropriate gene control responses. These riboswitches are primarily found in the 5’ untranslated regions of messenger RNAs. They are being divided into two parts- Evolutionary conserved sensor domain (an aptamer) which directly binds small molecules and an expression platform which undergoes structural changes in response to changes in aptamer [24]. Binding domains are highly conserved, even among divergent organisms. It is believed to have persisted through evolution. The Expression Platform Sequences vary widely among riboswitches, sequence even varies within a class. These parts of riboswitches in combination acts like the operon system which is based on the ON and OFF mechanism for the expression of the genes associated when the aptamer region is bound to the appropriate ligand to show the ligand’s efficiency onto the specific riboswitch binding site.
The most common mechanisms of riboswitches include: (1) formation of the hairpin structure that leads to RNA polymerase stalling and premature transcription termination, (2) base-pairing between Shine-Dalgarno and anti-Shine-Dalgarno sequence that blocks translation, and (3) changing the splice sites [25,26]. Riboswitches as a novel drug target and identify its potential inhibitor for H1N1, H2N2 and H3N2 strains of influenza virus.
BioRelix Inc. is building a club of anti-infective drug treatments that target riboswitches, or stretches of messenger RNAs that control genes expression and mechanism essential to the survival of many disease-causing microbes. This strategy has applications in the elimination of pathogens that are resistant to currently available drugs.
Material and Methods
The proposed framework for in-silico prediction and identification of riboswitches, their ligands and its inhibitors are given below:-
2.1 Identification of Gene Sequence for Riboswitch.
2.2 Human Nucleotide Database Search for Gene Sequence Similarity by BLAST
2.3 Transcription of Gene Sequence to Corresponding Riboswitch like Element (RLE).
2.4 Prediction of Structure of Riboswitch like Elements (RLE).
2.5 Blind Docking (BD).
2.6 Focused Docking (FD) for Identification of Ligand.
2.7 Refinement of Docking Results.
2.8 Virtual Screening Using NCI Diversity Set.
2.9 Evaluation of Physicochemical Properties of Lead Compounds.
2.10 Prediction of ADME & Toxicity of Lead Compounds.
The flow chart of framework is given in Fig. -1.
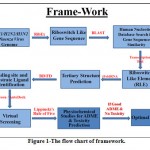 |
Figure 1: The flow chart of framework. |
Identification of Gene Sequence for Riboswitch –
The whole genomes of H1N1, H2N2 and H3N2 Influenza virus are obtained from the National Center for Biotechnology Information (NCBI) Gene Bank in FASTA format. The viral genome is searched for riboswitch like sequences present in it by online program Riboswitch Explorer to identify Riboswitch like sequences [6].
Human Nucleotide Database Search for Gene Sequence Similarity by BLAST
The viral riboswitch like gene sequences identified by RibEx are searched in the National Center for Biotechnology Information (NCBI) database for similar gene sequence present in Human (Homo Sapiens) genome by using Basic Linear Alignment Sequencing tool (BLAST).
 |
Figure 2: (A) Similarity of PRI1 gene sequence with Human genome using BLAST. (B) Similarity of PRI2 gene sequence with Human genome using BLAST. |
Transcription of Gene Sequence to Corresponding Riboswitch like Element (RLE)
The gene sequences identified by online program RibEx for riboswitch like elements are transcripted to its corresponding riboswitch by using online “Transcription and Translation Tool”.
Structure Prediction of Riboswitch
The 3-Dimensional structure of the riboswitch like elements is predicted by using online program iFoldRNA. iFoldRNA program performs interactive RNA folding simulations using discrete molecular dynamics simulations using coarse-grained structural RNA models to predict the 3d structure of the identified sequence of riboswitch like elements present in the genomes of H1N1, H2N2 and H3N2 Influenza virus.
Blind Docking (BD)
AutoDock based Blind docking (BD) technique is used to find out appropriate binding site present in the predicted riboswitch like structures of H1N1, H2N2 and H3N2 Influenza virus. In blind docking technique whole structure of riboswitch is covered under imaginary 3d grid box for docking. The coordinates of grid box used for blind docking technique for all the two identified riboswitches PRI1 and PRI2 are given in Table-1. [27,10,4,28]
Table 1: The coordinates of grid box for the Blind Docking (BD).
| Proteins | x-D | y-D | z-D | Spacing (Ả) | x center | y center | z center |
| PRI1 | 74 | 48 | 44 | 0.803 | -0.295 | 0.931 | 2.852 |
| PRI2 | 58 | 60 | 62 | 0.603 | 2.447 | -0.951 | 0.900 |
All computational studies are carried out using Autodock 4.2 installed on a single machine running on a 2.80 GHz Intel core2 duo processor with 3GB RAM and 320 GB hard disk with Windows XP as an operating system.
Docking software’s like “Autodock Tools”, “Autogrid” and “Autodock-4.2” were downloaded from the Scripps portal (http://autodock.scripps.edu). AutoDock software explores the whole surface of riboswitch for binding of ligand in the riboswitch. Alanine (ALA) is used as a ligand in blind docking technique to identify binding sites present in all the two predicted riboswitches, as it is the neutral and simplest ligand out of possible 20 amino acids as substrate of a riboswitch. Appropriate binding sites present in the surface of the predicted riboswitches are identified on the basis of lowest binding energy in the range of -5 to -15 Kcal/Mol. The appropriate binding site present in the predicted riboswitches of influenza is used for identification of binding residues involved in binding of ligand. The identified binding residues present in the predicted riboswitches are further utilized for focused docking.
Focused Docking (FD) for Identification of Ligand
Focused grid box covering ligand as well as binding residues involved in binding of ligand, is prepared for focused docking targeting specific ligand binding site for all the two identified riboswitches of Influenza virus [29-31]. The coordinates containing the information about the size and position of grid box used in focused docking of all the two predicted influenza riboswitches are tabulated in Table-2. These three grid boxes are further utilized for focused docking with 20 different amino acids for identification of specific substrate ligand for all the two predicted riboswitches.
Table 2: The coordinates of grid box for the Focussed Docking (FD).
| Proteins | x-D | y-D | z-D | Spacing (Ả) | x center | y center | z center |
| PRI1 | 40 | 30 | 36 | 0.303 | 14.085 | -8.772 | -9.892 |
| PRI2 | 46 | 38 | 44 | 0.308 | 3.623 | 0.654 | 7.768 |
Refinement of Docking Results
After identification of an appropriate ligand for predicted riboswitches by focused docking, separate docking of identified substrate ligand with its corresponding riboswitch is done for all the two riboswitches identified in the genome of Influenza virus repeatedly for a number of times to refine the results.
Virtual Screening Using NCI Diversity Set
The identified binding sites present in all the two predicted structures of novel Influenza riboswitches are utilized for virtual screening of NCI Diversity Set containing 1541 diverse molecules [32]. All the files necessary for virtual screening are prepared by software Raccoon. It is a graphical user interface for AutoDock virtual screening (autodock.scripps.edu/resources/raccoon). Raccoon can split multiple-molecule ligand library files, convert them into the AutoDock format (i.e. *. pdbqt), and filter them by using common criteria (e.g., Lipinski’s rules, fragment-like “rule of 3”, and drug-likeness). A validation check of the input files is performed at every step, which includes checking for the presence of non- standard atom types and ensuring that parameters, input file names, and grid maps have a coherent format.
Molecular docking simulation based virtual screening of all the two influenza riboswitches is done using similar docking and grid parameters used in focused docking earlier.
The coordinates of the grid box used in the virtual screening process of molecular libraries containing 1880 molecules against the binding site of riboswitches are tabulated in Table-3. The coordinates of grid box used in virtual screening of both the riboswitches PRI1 and PRI2 are given in Table-3.
Table 3: The coordinates of grid box for the Virtual Screening (VS).
| Proteins | x-D | y-D | z-D | Spacing (Ả) | x center | y center | z center |
| PRI1 | 40 | 30 | 36 | 0.303 | 14.085 | -8.772 | -9.892 |
| PRI2 | 46 | 38 | 44 | 0.308 | 3.623 | 0.654 | 7.768 |
Evaluation of Physicochemical Properties of Lead Compounds
The top five screened ligands for each riboswitch are evaluated for important physicochemical properties such as calculated partition coefficient (ClogP), 2D-Polar surface area (2D PSA), molecular weight, hydrogen bond donor and acceptor sites etc. by using Marvin Sketch software.
Prediction of ADME and Toxicity of Lead Compounds
The top 5 lead molecules for both the two riboswitches are evaluated by using the OSIRIS online program for toxicity and ADME properties [33]. This program evaluates the presence of major toxicities such as mutagenicity, tumorigenicity, irritant effect and reproductive effects in the lead molecules on the basis of functional group present in their chemical structure. This program also calculates drug-likeness and drug score of the lead molecules on the basis of their physicochemical properties.
The results obtained in each of the above steps are presented in the next section.
Results and Discussion
The stepwise results obtained are summarized and discussed below:
Identified Genes for Riboswitch like Sequence
The following two gene sequences are identified by the RibEx for transcription into riboswitch like elements present in the viral DNA of H1N1, H2N2 and H3N2 Influenza virus genome.
PRI1- TATGAGGCCCATACAACTGGCAAGTGCACCAGCAGAATAA
PRI2- ATCCCAAAATCCCCTTAGTCAGAGG
Gene Sequence Similarity with Human Genome
The identified riboswitch like gene sequences of H1N1, H2N2 and H3N2 Influenza Virus shows the following results on the NCBI nucleotide database search for gene sequence similarity present in human (Homo sapiens) genome.
(a) PRI1- The gene sequence of PRI1 riboswitch shows a BLAST score 38.2 with a maximum 97% of gene sequence similarity present in the human genome. The BLAST result of gene sequence similarity to PRI1 gene sequence is shown in Fig.-2(A). The human genome showing 38.2 blast score is searched for the presence of any riboswitch like gene sequence by using RibEx software. No riboswitch was found in similar gene in the human genome.
(b) PRI2- The gene sequence of SPR1 riboswitch shows a BLAST score 34.2 with a maximum 72% gene sequence similarity present in the human genome. The BLAST result of gene sequence similarity to PRI2 gene sequence is shown in Fig-2(B). The human genome showing 34.2 blast score is searched for the presence of any riboswitch like gene sequence by using RibEx software. No riboswitch was found in similar gene in the human genome.
Transcription of Gene Sequence to Riboswitch like Element Sequence
Following sequences are identified after transcription of identified gene sequences of Riboswitch like elements;
PRI1- UAUGAGGCCCAUACAACUGGCAAGUGCACCAGCAGAAUAA
PRI2- AUCCCAAAAUCCCCUUAGUCAGAGG
The identified two DNA gene sequences are transcripted to corresponding riboswitch like elements using online, “Transcription and Translation Tool”. The tool replaces the thymine (T) nucleotide of the gene sequence with uracil (U) to form its corresponding riboswitch (mRNA).
Predicted tertiary Structures of Riboswitch
The tertiary structures of both the identified riboswitches in H1N1, H2N2 and H3N2 Influenza virus are predicted by online program iFoldRNA, and following results are obtained.
(a) The tertiary structure of Influenza riboswitch PRI1 is identified by using online program iFoldRNA. The PRI1 riboswitch consists of a single chain of 40 nucleotides. The predicted tertiary structure of PRI1 riboswitch in Influenza virus is shown in Fig. 3.
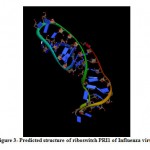 |
Figure 3: Predicted structure of riboswitch PRI1 of Influenza virus. |
(b) The tertiary structure of the Influenza riboswitch PRI2 is identified by using online program iFoldRNA. The PRI2 riboswitch consists of a single chain of 25 nucleotides. The predicted tertiary structure of PRI2 riboswitch in Influenza bacteria is shown in Fig. 4.
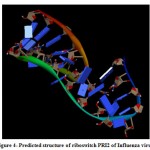 |
Figure 4: Predicted structure of riboswitch PRI2 of Influenza virus. |
Blind and Focused Docking Results
(a) The binding site present in the Influenza riboswitch PRI1 identified by Blind docking suggests that the residues ADE34, ADE31 CYT33, GUA24 and GUA32 of PRI1 riboswitch are involved in the binding of ligand.
The focused docking of PRI1 riboswitch using the identified binding residues involved in the binding of the ligand with the riboswitch suggests the Lysine as its substrate ligand as it shows best binding among 20 amino acids with a binding energy value of -7.26 Kcal/Mol (Ki = 4.74 μM). The ligand binding site identified in predicted three dimensional structure of PRI1 riboswitch of Influenza virus is shown in Fig. -6.
 |
Figure 5: Grid box used for focused docking of PRI1 riboswitch covering all binding residues involved in binding of ligand. |
 |
Figure 6: Grid box used for focused docking of PRI2 riboswitch covering all binding residues involved in binding of ligand. |
The focused docking result of PRI1 riboswitch is tabulated in Table-4.
Table 4: Focused Docking Results for PRI1
| Amino acid | Binding Energy (Kcal/Mol) | Ki ( M) |
| 1. ALA | -5.24 | 144.21 |
| 2. ARG | -6.22 | 27.39 |
| 3. ASN | -5.61 | 77.27 |
| 4. ASP | -5.5 | 93.13 |
| 5. CYS | -5.15 | 166.65 |
| 6. GLN | -6.18 | 29.62 |
| 7. GLU | -6.12 | 32.44 |
| 8. GLY | -5.06 | 196.66 |
| 9. HIS | -5.02 | 207.94 |
| 10. ILE | -5.68 | 68.27 |
| 11. LEU | -5.8 | 55.72 |
| 12. LYS | -7.26 | 4.74 |
| 13. MET | -5.21 | 151.69 |
| 14. PHE | -5.29 | 132.87 |
| 15. PRO | -4.95 | 237.24 |
| 16. SER | -5.69 | 67.11 |
| 17. THR | -5.97 | 41.82 |
| 18. TRP | -5.57 | 82.57 |
| 19. TYR | -5.21 | 151.98 |
| 20. VAL | -5.9 | 47.45 |
The binding site present in the Influenza riboswitch PRI2 is identified by blind docking suggests that the residues CYT11, URI10, ADE21, GUA22 and ADE23 of PRI2 riboswitch are involved in the binding of ligand.
The focused docking of PRI2 riboswitch using the identified binding residues involved in the binding of the ligand with the riboswitch suggests the Tryptophan as its substrate ligand as it shows best binding among 20 amino acids with a binding energy value of -6.77 Kcal/Mol (Ki = 10.91μM). The ligand binding site identified in predicted three dimensional structure of PRI2 riboswitch of Influenza virus is shown in Fig.-7.
 |
Figure 7: Toxicity and drug likeness prediction of Lead Compound NCI_81462_a. |
The focused docking result of PRI2 riboswitch is tabulated in Table-5.
Table 5: Focused Docking Results for PRI2
| Amino acid | Binding Energy (Kcal/Mol) | Ki ( M) |
| 1. ALA | -5.05 | 199.83 |
| 2. ARG | -6.37 | 21.28 |
| 3. ASN | -6.23 | 27.08 |
| 4. ASP | -3.7 | 1940 |
| 5. CYS | -5.29 | 133.44 |
| 6. GLN | -5.61 | 77.41 |
| 7. GLU | -4.2 | 840.94 |
| 8. GLY | -4.88 | 264.43 |
| 9. HIS | -6.13 | 32.32 |
| 10. ILE | -5.64 | 72.83 |
| 11. LEU | -5.69 | 67.4 |
| 12. LYS | -6.33 | 23.02 |
| 13. MET | -5.37 | 115.58 |
| 14. PHE | -6.11 | 33.07 |
| 15. PRO | -4.66 | 382.37 |
| 16. SER | -5.35 | 119.04 |
| 17. THR | -5.6 | 78.58 |
| 18. TRP | -6.77 | 10.91 |
| 19. TYR | -6.19 | 29.22 |
| 20. VAL | -5.44 | 103.33 |
Docking results obtained by blind docking of both the riboswitches i.e. PRI1 and PRI2 are tabulated in Table-6.
Table 6: Blind Docking Results
| S. No. | Ligand | Binding Energy (Kcal/Mol) | Ki (μM) |
| PRI1 | ALA | -5.24 | 144.21 |
| PRI2 | ALA | -5.05 | 199.83 |
Refined Docking Results
The results of individual docking of riboswitch PRI1 with Lysine for a repeated number of times are given in Table-7. There was a change in its binding energy from -7.26 to – Kcal/Mol.
Similarly the Arginine is repetitively docked with PRI2 for refinement of their binding energy values. The binding energy for PRI2 riboswitch is refined from -6.37 to – Kcal/Mol. These refined binding values are tabulated in Table-7.
Table 7: Refined Docking Results
| S. No. | Riboswitch | Ligand | Binding Energy (Kcal/Mol) | Ki (μM) |
| 1 | PRI1 | ALA | ||
| 2 | PRI2 | ALA |
Virtual Screening Results
The 5 lead molecules for each riboswitch were selected after virtual screening of NCI Diversity Set containing 1541 diverse molecules against all the three predicted Influenza riboswitches PRI1 and PRI2. The binding energy and Ki value of 5 lead molecules for both of the influenza riboswitches are given in Table-8 and 9 for each of the predicted riboswitch PRI1 and PRI2 respectively.
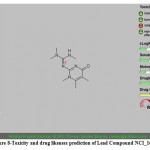 |
Figure 8: Toxicity and drug likeness prediction of Lead Compound NCI_168184. |
(a) The following five lead molecules were obtained for PRI1 riboswitch after the virtual screening: NCI_81462_a, NCI_168184, NCI_113486, NCI_207895 and NCI_3076_a. The binding energy and Ki value of top 5 lead molecules for PRI1 riboswitches is given in Table-8.
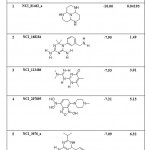 |
Table 8: List of Binding Energies of Proposed lead molecules for PRI1 |
(b) The five lead molecules were obtained for PRI2 riboswitch after the virtual screening: NCI_117268_a, NCI_170621, NCI_281816_a, NCI_403379 and NCI_13316_b. The binding energy and Ki value of top 5 lead molecules for PRI1 riboswitches is given in Table-9.
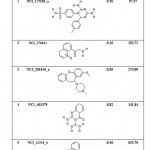 |
Table 9: List of Binding Energies of Proposed lead molecules for PRI2 |
Physicochemical Properties of Screened Lead Compounds
(a) The physicochemical properties of screened top 5 lead molecules for PRI1 influenza riboswitch are tabulated in Table-10.
(b) The physicochemical properties of screened top 5 lead molecules for PRI2 Influenza riboswitch is tabulated in Table-11.
Table 10: Physiochemical Parameters of Proposed lead molecules for PRI1 Riboswitch.
| Parameter | Optimum range | NCI_81462_a | NCI_168184 | NCI_113486 | NCI_207895 | NCI_3076_a |
| ClogP | -5 to +5 | 0.37 | 0.22 | -0.21 | -2.26 | 1.44 |
| PSA (2D) (Å2) | 60 to 140 | 39.33 | 106.02 | 80.58 | 101 | 50.39 |
| Mol. Wt. | 150 to 500 | 182 | 246 | 209.24 | 281 | 293 |
| HBD | 0 to 5 | 3 | 4 | 2 | 4 | 2 |
| HBA | 0 to 10 | 4 | 4 | 5 | 8 | 4 |
Table 11: Physiochemical Parameters of Proposed lead molecules for PRI2 Riboswitch.
| Parameter | Optimum range | NCI_117268_a | NCI_170621 | NCI_281816_a | NCI_403379 |
NCI_13316_b |
| ClogP | -5 to +5 | 1.68 | 0.71 | 4.68 | -0.65 | 4.56 |
| PSA (2D) (Å2) | 60 to 140 | 140 | 63.17 | 6.48 | 108 | 45.15 |
| Mol. Wt. | 150 to 500 | 358 | 203 | 356 | 205 | 346 |
| HBD | 0 to 5 | 3 | 1 | 1 | 3 | 2 |
| HBA | 0 to 10 | 7 | 3 | 1 | 5 | 2 |
ADME and Toxicity Profiling
The results of toxicity and ADME prediction of screened top 5 lead compounds for both the riboswitches is computed by using Osiris online program.
(a) The toxicity prediction and drug likeness score for top 5 virtually screened lead molecule for PRI1 riboswitch are shown in Fig. 7 to 11. The three lead compounds out of top 5 screened leads ZINC19325791, ZINC19362650 and ZINC08652230 passed the toxicity test with a good drug likeness score, while lead ZINC01556940 shows satisfactory drug score but there is a probability of presence of some reproductive effects is shown in the toxicity test. The lead compound ZINC19230120 shows the poor drug likeness score and supposed to be having serious toxic effects such as mutagenicity, irritant nature and reproductive effects.
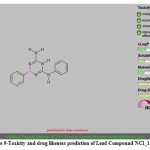 |
Figure 9: Toxicity and drug likeness prediction of Lead Compound NCI_113486. |
 |
Figure 10: Toxicity and drug likeness prediction of Lead Compound NCI_207895. |
 |
Figure 11: Toxicity and drug likeness prediction of Lead Compound NCI_3076_a. |
The toxicity prediction and drug likeness score for top 5 virtually screened lead molecule for PRI2 riboswitch are shown in Fig. 12 to 16. All the five screened lead compounds ZINC01584497, ZINC03947435, ZINC13597738, ZINC01729525 and ZINC01871223 passed the toxicity test without any toxic effect and satisfactory drug likeness score.
 |
Figure 12: Toxicity and drug likeness prediction of Lead Compound NCI_117268_a. |
 |
Figure 13: Toxicity and drug likeness prediction of Lead Compound NCI_170621. |
 |
Figure 14: Toxicity and drug likeness prediction of Lead Compound NCI_281816_a. |
 |
Figure 15: Toxicity and drug likeness prediction of Lead Compound NCI_403379. |
 |
Figure 16: Toxicity and drug likeness prediction of Lead Compound NCI_13316_b. |
Conclusion
We have been able to perform in-silico prediction of two riboswitches for H1N1, H2N2 and H3N2 strains of Influenza A. The in-silico approach has further been successfully used to predict binding sites and specific ligands for these strains of influenza and lead molecules which inhibit these riboswitches. In all the three strains of influenza Lysine is found to be the most efficient ligand.
Thus, we conclude that in-silico approach can be used for prediction of riboswitches, their binding sites, specific ligands, and their potential inhibitors.
The inhibitors predicted are free from the side effects of the antiviral drugs and have remote chances of developing resistance. In all such studies can be useful for developing novel drugs for infectious disease like influenza.
Acknowledgments
Authors acknowledge Department of Mathematics, Bioinformatics & Computer Application, Maulana Azad National Institute of Technology (M.A.N.I.T.), Bhopal, for providing the facilities to carry out the project under the immense guidance of Dr. K.R. Pardasani (Professor, Department of Mathematics and Former Head of Department of Mathematics, Bioinformatics & Computer Application).
References
- Somdutt M, Kamal RP (2012) Riboswitch as a target for Streptococcus pneumoniae. ONLINE J BIOINFORMATICS 13(2):285-313
- Hall CB (2007) The spread of influenza and other respiratory viruses: complexities and conjectures. CLIN INFECT DIS 45 (3):353-359. doi:10.1086/519433
- Brankston G, Gitterman L, Hirji Z, Lemieux C, Gardam M (2007) Transmission of influenza A in human beings. Lancet Infect Dis 7 (4):257-265. doi:10.1016/S1473-3099(07)70029-4
- Tellier R (2006) Review of aerosol transmission of influenza A virus. Emerg Infect Dis 12 (11):1657
- Bell D (2006) Non-pharmaceutical interventions for pandemic influenza, international measures. EMERG INFECT DIS 12 (1):81-87. doi:10.3201/eid1201.051370
- KLONTZ KC, HYNES NA, GUNN RA, WILDER MH, HARMON MW, KENDAL AP (1989) An outbreak of influenza A/Taiwan/1/86 (H1N1) infections at a naval base and its association with airplane travel. AM J EPIDEMIOL 129 (2):341-348
- Moser MR, Bender TR, Margolis HS, Noble GR, Kendal AP, Ritter DG (1979) An outbreak of influenza aboard a commercial airliner. AM J EPIDEMIOL 110 (1):1-6
- Glezen WP, Couch RB, MacLean RA, Payne A, Baird JN, Vallbona C, Tristan M, Byrd N (1978) Interpandemic influenza in the Houston area, 1974–76. NEW ENGL J MED 298 (11):587-592. doi:10.1056/NEJM197803162981103
- MONTO AS, KIOUMEHR F (1975) The Tecumseh study of respiratory illness IX. Occurrence of influenza in the community, 1966–1971. AM J EPIDEMIOL 102 (6):553-563
- Walsh EE, Cox C, Falsey AR (2002) Clinical features of influenza A virus infection in older hospitalized persons. J AM GERIATR SOC 50 (9):1498-1503. doi:10.1046/j.1532-5415.2002.50404.x
- Keren R, Zaoutis TE, Bridges CB, Herrera G, Watson BM, Wheeler AB, Licht DJ, Luan XQ, Coffin SE (2005) Neurological and neuromuscular disease as a risk factor for respiratory failure in children hospitalized with influenza infection. JAMA-J AM MED ASSOC 294 (17):2188-2194. doi:10.1001/jama.294.17.2188
- Bright RA, Medina M-j, Xu X, Perez-Oronoz G, Wallis TR, Davis XM, Povinelli L, Cox NJ, Klimov AI (2005) Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. LANCET 366 (9492):1175-1181. doi:10.1016/s0140-6736(05)67338-2
- Updated Interim Recommendations for the Use of Antiviral Medications in the Treatment and Prevention of Influenza for the 2009-2010 Season. (2009). http://www.cdc.gov/h1n1flu/recommendations.htm.
- Bautista E, Chotpitayasunondh T, Gao Z, Harper SA, Shaw M, Uyeki TM, Zaki SR, Hayden FG, Hui DS, Kettner JD (2010) Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. doi:10.1056/NEJMra1000449
- Harper SA, Bradley JS, Englund JA, File TM, Gravenstein S, Hayden FG, McGeer AJ, Neuzil KM, Pavia AT, Tapper ML (2009) Seasonal influenza in adults and children—diagnosis, treatment, chemoprophylaxis, and institutional outbreak management: clinical practice guidelines of the Infectious Diseases Society of America. CLIN INFECT DIS 48 (8):1003-1032. doi:10.1086/598513
- Gubareva LV, Matrosovich MN, Brenner MK, Bethell RC, Webster RG (1998) Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J INFECT DIS 178 (5):1257-1262. doi:10.1086/314440
- Stephenson I, Democratis J, Lackenby A, McNally T, Smith J, Pareek M, Ellis J, Bermingham A, Nicholson K, Zambon M (2009) Neuraminidase inhibitor resistance after oseltamivir treatment of acute influenza A and B in children. CLIN INFECT DIS 48 (4):389-396. doi:10.1086/596311
- Gubareva LV, Kaiser L, Matrosovich MN, Soo-Hoo Y, Hayden FG (2001) Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J INFECT DIS 183 (4):523-531. doi:10.1110/ps.0202302
- Barnett J, Cadman A, Gor D, Dempsey M, Walters M, Candlin A, Tisdale M, Morley P, Owens I, Fenton R (2000) Zanamivir susceptibility monitoring and characterization of influenza virus clinical isolates obtained during phase II clinical efficacy studies. ANTIMICROB AGENTS CH 44 (1):78-87. doi:10.1128/AAC.44.1.78-87.2000
- Kiso M, Mitamura K, Sakai-Tagawa Y, Shiraishi K, Kawakami C, Kimura K, Hayden FG, Sugaya N, Kawaoka Y (2004) Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364 (9436):759-765. doi:10.1016/s0140-6736(04)16934-1
- Ison MG, Gubareva LV, Atmar RL, Treanor J, Hayden FG (2006) Recovery of drug-resistant influenza virus from immunocompromised patients: a case series. J INFECT DIS 193 (6):760-764. doi:10.1086/500465
- Farias JA, Fernández A, Monteverde E, Vidal N, Arias P, Montes MJ, Rodríguez G, Allasia M, Ratto ME, Jaén R (2010) Critically ill infants and children with influenza A (H1N1) in pediatric intensive care units in Argentina. INTENS CARE MED 36 (6):1015-1022. doi:10.1007/s00134-010-1853-1
- Karie S, Launay-Vacher V, Janus N, Izzedine H, Deray G (2006) Pharmacokinetics and dosage adjustment of oseltamivir and zanamivir in patients with renal failure. NEPHROL DIAL TRANSPL 21 (12):3606-3608. doi:10.1093/ndt/gfl345
- Mandal M, Breaker RR (2004) Gene regulation by riboswitches. NAT REV MOL CELL BIO 5 (6):451-463. doi:10.1038/nrm1403
- Cheah MT, Wachter A, Sudarsan N, Breaker RR (2007) Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 447 (7143):497-500. doi:10.1038/nature05769
- Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR (2007) Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell 19 (11):3437-3450
- Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR (2006) Antibacterial lysine analogs that target lysine riboswitches. NAT CHEM BIOL 3 (1):44-49. doi:10.1038/nchembio842
- Baird NJ, Kulshina N, D’Amaré ARF (2010) Riboswitch function: flipping the switch or tuning the dimmer? RNA BIOL 7 (3):328-332. doi:10.4161/rna.7.3.11932
- Iorga B, Herlem D, Barré E, Guillou C (2006) Acetylcholine nicotinic receptors: finding the putative binding site of allosteric modulators using the “blind docking” approach. J MOL MODEL 12 (3):366-372. doi:10.1007/s00894-005-0057-z
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J COMPUT CHEM 30 (16):2785-2791. doi:10.1002/jcc.21256
- Famulok M, Hartig JS, Mayer G (2007) Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. CHEM REV 107 (9):3715-3743. doi:10.1021/cr0306743
- Bikádi Z, Hazai E, Zsila F, Lockwood SF (2006) Molecular modeling of non-covalent binding of homochiral (3< i> S</i>, 3′< i> S</i>)-astaxanthin to matrix metalloproteinase-13 (MMP-13). BIOORGAN MED CHEM 14 (16):5451-5458. doi:10.1016/j.bmc.2006.04.047
- Hetényi C, van der Spoel D (2002) Efficient docking of peptides to proteins without prior knowledge of the binding site. PROTEIN SCI 11 (7):1729-1737. doi:10.1110/ps.0202302

This work is licensed under a Creative Commons Attribution 4.0 International License.





