Manuscript accepted on :
Published online on: 12-01-2016
Industrial Important Enzymes from Alkaliphiles – An Overview
Arunachalam Chinnathambi*
Department of Botany and Microbiology, College of Science, King Saud University, Riyadh-11451, Kingdom of Saudi Arabia
DOI : http://dx.doi.org/10.13005/bbra/1868
ABSTRACT: Alkaliphiles are an interesting group of extremophilic organisms that thrive at pH of 9.0 and above and which are represented by prokaryotes, eukaryotes, and archaea. It is clear that a variety of taxa are represented among the alkaliphiles, some of which are new. Alkaliphiles can be isolated from normal environments such as garden soil, although the number isolated increases dramatically in highly alkaline environments. Many alkaliphile-products, notably enzymes, have found widespread practical applications, primarily in the detergent and laundry industries; other potentially important products isolated from alkaliphiles include antibiotics and carotenoids. Alkaliphiles, and their products, are also potentially important for use in the degradation of xenobiotics, and they play a major role in the biogeocycling of key inorganic compounds. This review provides an insight into the huge diversity of alkaliphilic bacteria, the varied products obtained from them, and the need for further investigations on this interesting group of bacteria.
KEYWORDS: Alkaliphiles; Enzyme; Detergents; Food industry
Download this article as:| Copy the following to cite this article: Chinnathambi A. Industrial Important Enzymes from Alkaliphiles – An Overview. Biosci Biotechnol Res Asia 2015;12(3) |
| Copy the following to cite this URL: Chinnathambi A. Industrial Important Enzymes from Alkaliphiles – An Overview. Biosci Biotechnol Res Asia 2015;12(3). Available from: https://www.biotech-asia.org/?p=3705 |
Introduction
The majority of microorganisms occur, both in terms of numbers and diversity, in what can be termed “moderate” environments. Microbes also occur generally with less diversity and in lower numbers in so-called “extreme” environments on Earth in which life was previously thought to be restricted or absent prevent the existence of life. Extremophilic microorganisms exhibit the ability to grow at the limits of a variety of environmental factors, which critically influence growth, such as pH, salinity, temperature and pressure; the organisms which grow in such environments are respectively called alkaliphiles, halophiles, thermophiles and acidophiles, 1, 2
Since, alkaline enzymes play a major role in the global enzyme market, it is obviously important that they and the microbes which produce them bee intensively studied. Enzymes obtained from alkaliphiles are stable when added to detergents because of their inherent tolerance to high pH; they can also generally function in the presence of bleach. Enzyme–based detergents can also generally function at lower temperatures that equivalent chemical detergents and are generally available at lower cost. Put simply, they are generally cheaper and not require high temperatures to provide efficient washing 2
Alkaliphilic microorganisms are found not only found in environments having neutral or high pH but can also be isolated from acidic soils 3, probably because soils having areas which have a bulk pH which is neutral or acidic also possess localised alkaline pockets in which alkaliphiles can thrive. Enzymes from these microorganisms have found major commercial applications such as providing additives to laundry detergents, for use in efficient food processing, in the finishing of fabrics, and for use in the pulp and paper industries.
Here, we will highlight the main enzymes which are derived from alkaliphiles and discuss their industrial application.
Definition – Alkaliphiles
A number of microorganisms exhibit more than one pH optimum for growth depending on the prevailing growth conditions, particularly in relation to nutrients, metal ions and temperature. Alkaliphiles are microorganisms that grow well, or optimally at pH values above 9, often between 10 and 12, but do not grow at the near neutral pH of 6.5 3 (Figure 1)
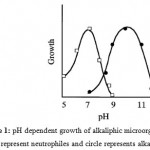 |
Figure 1: pH dependent growth of alkaliphic microorganism. Squares represent neutrophiles and circle represents alkaliphiles3. |
Background of the Study
Few papers on alkaliphiles were published during in the late nineteen sixties, but since then interest in this group, particularly in relation to their ability to produce industrially useful enzymes has burgeoned. The first paper reporting the production of a alkaline enzyme by an alkaliphilic microorganism appeared in 19714. Traditionally, the Japanese used indigo to dye cloth which had been naturally reduced under alkaline conditions in the presence of slaked lime, wood ash and Japanese sake. Indigo derived from indigo leaves is reduced by particular bacteria that grow under these highly alkaline conditions in a traditional process called “indigo fermentation”. The most important factor in this process is pH control which was formerly controlled by the skill of craftsmen.
Alkaliphiles consist of two main physiological groups of microorganisms; alkaliphiles and haloalkaliphiles. Alkaliphiles require an alkaline pH of 9 or more for their growth and have an optimal growth pH of around 105. Haloalkaliphiles, in contrast, require both an alkaline pH (pH 9) and high salinity (up to 33% NaCl). Alkaliphiles have been isolated mainly from neutral environments, sometimes even from acidic soil samples and feces. Haloalkaliphiles are mainly found in extremely alkaline saline environments, such as the Rift Valley lakes of East Africa and the western soda lakes of the United States6,7.
Distribution of Alkaliphiles
Alkaliphiles have a world-wide distribution in, deep sea, alkaline lake, sub-ground soils and etc. They include, amongst other bacterial groups, aerobic spore-formers, anaerobic non-spore-formers, halophiles, thermophiles, archaea, psychrophiles and piezophile; alkaliphiles fungi, yeast and phages have also been isolated.
Naturally occurring alkaline environments, such as carbonate springs, alkaline soils, and soda lakes, are characterized by their high values (pH 8.0–11.0) due to the presence of high concentrations of sodium carbonate salts which are generally formed by evaporative concentration8. The Egyptian hyper saline soda lakes in the Wadi Natrun area (30°15’N, 30°30’E) are an excellent example of these hitherto unexplored alkaline ecosystems. The following figure shows the relationship between soil pH and the distribution of alkaliphiles (Figure 2).
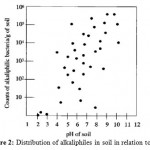 |
Figure 2: Distribution of alkaliphiles in soil in relation to pH. |
Types of alkaliphiles
Alkaline-adapted micro-organisms can be classified into two main groupings, alkaliphiles (also called alkalophiles) and alkalitolerant species. The term alkaliphiles (alcali from Arabic, soda ash, phile, loving) is generally restricted to those micro-organisms that actually require alkaline conditions for growth and the optimum growth rate of these microorganisms is observed in at least two pH units above neutrality. Organisms capable of growing at pH values more than 9 or 10 but, since they exhibit optimum growth rates at around neutrality or less, these are referred to as alkalitolerant9,10 .
Aerobic alkaliphiles
Alkaliphilic microorganisms can grow in neutrophilic conditions, as well as occupying specific extreme environments in nature. The medium used to isolate alkaliphilic microorganism must, obviously, be alkaline and usually sodium carbonate is added to adjust the pH to around 10. The population of such microorganisms in soil can reach between 102 to 105 CFU/ml of soil, this value corresponding to around 1/10 to 1/100 of the population of the neutrophilic microorganisms1 . Recent studies show that alkaliphilic bacteria have also been found in deep-sea sediments collected from depths up to the 10,898 m of the Mariana Trench11 .
Anaerobic Alkaliphiles
Wiegel12 has provided a comprehensive review of the isolation, diversity and phylogeny of anaerobic alkaliphiles microbes. Niimura13 was first reported the occurrence of anaerobic alkaliphilic microorganism, although though no taxonomic details were initially reported. Subsequently, many anaerobic spore-forming alkaliphiles have been isolated using conventional procedures and their enzymes studied. Nine moderately alkalitotolerant thermophilic bacteria, possessing similar properties, have also been isolated from water and soil samples obtained from Yellowstone National Park, including strain JW/YL-138T and eight similar strains that represent a new genus and species, namely, Anaerobranca horikoshii. At 60°C the pH range for growth of this bacterium is 6.9 to 10.3, with an optimum at pH 8.5. The pH range for growth was found to be between 6.5 to 10.5, with an optimum around 9.0 and both polysulfide and elemental sulfur were reduced to H2S.
Haloalkaliphiles
Halkohalophiles occur in alkaline environments such as soda desert and soda lakes, examples of which are the extremely alkaline lakes found in Kenya (Magadi) and Egypt (Wadi Natrun) which contains the alkalinity of pH 10.5 to 12. Hypersaline soda lakes are particularly rich in populated by alkaliphilic representatives of halophilic archaea. McGenity14 isolated a novel haloalkaliphilic archaea from Lake Magadi. Cells of this organism contain large gas vacuoles in the stationary phase of growth, and colonies produced by these archaea are bright pink; the name Natronobacterium vacuolatasp. nov. has been proposed for this organism. Many organisms isolated from alkaline and highly saline environments, such as soda lakes, also require high salinity, which is achieved by adding NaCl to the isolation medium. Kamekura15 studied the diversity of alkaliphilic halobacteria on the basis of phylogenetic tree reconstructions, signature bases specific for individual genera, and sequences of spacer regions between 16S and 23S rRNA genes. They proposed the following changes: Natronobacterium pharaonisto to be transferred to Natronomonasgen. nov. as Natronomonas pharaonis gen. nov. comb. nov.; Natronobacterium vacaolatum to be transferred to the genus Halorubrum as Halorubrum vacuolatum comb. nov.; and Natronobacterium magadii to be transferred to the genus Natrialba as Natrialba magadii.
The isolated two haloalkaliphilic archaea from a soda lake in Tibet16. On 16S rRNA phylogenetic trees, the two strains formed a monophyletic cluster. They differed from their closest neighbors, Halobacterium trapanicum and Natrialba asiatica, in polar lipid composition, as well as physiological and phenotypic characteristics, the strains should be classified in a new genus, Natronorubrum gen. nov. Other common osmolytes (glycine, betaine, glutamate, and proline) were found not to be accumulated or used for osmotic balance in place of the sulfotrehalose by these halophilic archaea17. They belong to the genus Clostridia and the Deltaproteobacteria, respectively, and represent moderate salt-tolerant non sulfate-reducing alkaliphiles.
Enzyme Production
Advances in the application of alkaliphilic or alkali tolerant based biomolecules during the past 20 years are due in the main to the introduction to the detergent industry of proteolytic enzymes classified as serine protease. Since the discovery of this enzyme in the 1970s, attention has been centred on alkaliphilic enzymes so that within a few years a large number of enzymes have become available, including alkaline proteases, amylases, pectinases, pullulanase, cellulase, alginases, catalase, RNase, DNase, restriction enzyme, β1,3-glucanase, xylanase, α-galactosidase, β-galactosidase, penicillinase, maltose dehydrogenase, glucose dehydrogenase, uricase, polyamine oxidase, β-mannanase, and β-mannosidase18.
Protease
Proteases constitute a very important group of industrial enzymes whose global sales amount to the order of 60% of the total enzyme market, of which alkaline proteases constitute 25%19. Horikoshiin 1991 first documented that Bacillus sp. strain 221 was capable of secreting alkaline serine protease and since then, many alkaline proteases from other Bacillus sp. have been extensively studied, characterized, and commercialized20. ‘‘Burnus,’’ was launched in 1913 and was the first enzymatic detergent made of sodium carbonate and pancreatic extract of trypsin, while the first detergent preparation containing bacterial enzyme (BIO-40) was marketed in 1956. The isolated a new alkaline protease from an alkaliphilic Bacillus sp21 and Kwon22 isolated alkaline proteases, named VapT and VapK, from Gram-negative alkaliphilic Vibrio metschnikovii strain RH530. Both enzymes have pH and temperature optima of 10.5 and 60°C, respectively. Han and Damodaran23 reported the purification and characterization of an extracellular endopeptidase from Bacillus pumilus displaying high stability in 10% (wt/vol) sodium dodecyl sulfate and 8 M urea. The genes coding for alkaline protease, stable in organic solvent were cloned in Pseudomonas aeruginosa strain K, and Verma24 studied alkaline protease production by Thermoactinomyces sp. RS1 when grown on locally available inexpensive agricultural and household wastes.
Industrial application
Protease plays a major role in industrial applications of, for example: detergent formulations, contact lens solutions, cheese and meat product processing, dehairing and for the recovery of silver from photographic films25,19,26. The literature on the application of industrially important protease enzymes has been reviewed in detailed by Anwar and Saleemuddin27 and Kumar and Takagi28 a list of commercially produced alkaline proteases used in detergent formulations, silk degumming, food and feed industry, photographic gelatin hydrolysis, leather dehairing, cosmetics, and pharmaceuticals can be found in the comprehensive26.
Detergents
Some enzymes produced by microorganisms are now commercially available as detergent additives. Alkaline serine proteases, active at pH 7.0 – 11.0 for example, are key components of laundry detergents26. It is desirable that proteases maintain their activity in the presence of other constituents making up the detergent formulation such as surfactants19, 26.
Dehairing
The conventional dehairing techniques used in the leather industry involve the use of harmful and polluting chemicals such as sodium sulphide. Keratinolytic enzymes can replace such chemical approaches and lead to the production of a higher quality hide and a reduction in the production of toxic wastes. In this process, the non-collagenous hide components are subjected to protease-mediated hydrolysis which results in more rapid water absorption, a reduced soaking time and a reduction on the production of environmental pollutants. Keratinases are proteolytic enzymes that can break down keratin-based substrates. Most keratinases are serine or metalloproteases and function best at vpH ranging from neutral to alkaline and temperatures of 40–60°C Jaouadi29 have also reported that an extracellular alkaline rSAPB protease from B. subtilis DB430/pNZ1 provided an ecofriendly method of producing goat hides which completely eliminates the use of polluting lime and sulphides.
Amylases
Although alkaline amylases produced by neutrophilic microorganisms have yet to be reported, an alkaline amylase was produced in Horikoshi-II medium by the alkaliphilic Bacillus sp. No. A-40-24. Alkaline amylases are classified into four types according to their pH activity curves. The type-I curve has only one peak at pH 10.5; the type-II curve has two peaks at pH 4.0-4.5 and 9.0-10.0; the type-III curve has three peaks at pH 4.5, 7.0 and 9.5-10.0; finally, the type-IV curve has one peak at pH 4.0 with a shoulder at pH 10.0. Type-Ill amylase (Bacillus sp. No. 38-2 enzyme) and type-IV amylase (Bacillus sp. No. 17 and Bacillus sp. No.13 enzymes) exhibit high cyclomaltodextrin glucanotransferase activity which converts starch to cyclodextrins30. Alpha-amylases act on the a-1,4 bonds between adjoining glucose units leading to the formation of glucose, maltose, and maltotriose.
Kim31 studied that the alkaliphilic Bacillus sp. strain GM8901 which produces five alkaline amylases in culture. McTigue32 also reported that alkaline amylases of three alkaliphilic Bacillus strains. Bacillus halodurans A-59 (ATCC 21591), Bacillus sp. strain NCIB 11203 and Bacillus sp. strain IMD370 produced alkaline α-amylases with maximum activity at pH 10.0, and Kelly33 found that the alkaline amylase of Bacillus sp. strain IMD370 could hydrolyze raw starch.
Several attempts have been made to produce cyclodextrins these unique compounds on an industrial scale. In 1969, Corn Products International Co. produced /β-CD using B. macerans CGTase. Teijin Ltd of Japan also produced /β-CD using the B. macerans enzyme in a pilot plant, although the following serious problems were encountered in both production processes: (1) CGTase from B. macerans is not suitable for industrial use because the enzyme is not thermostable enough. (2) yields of CD from starch are not high, usually 20% to 30% on an industrial scale, and (3) toxic organic solvents such as trichloroethylene, bromobenzene, toluene, etc. have to be used to precipitate CD due to low conversion rates. A CGTase produced by the alkaliphilic Bacillus sp. No. 38-2 overcame all these weak points and was used to mass produce crystalline CD’s at low cost without the use of organic solvents. The yield of CD ranged from 85 to 90% for amylose to 70 to 80% for potato starch used on a laboratory scale. Due to the high conversion rate, CDs can be directly crystallized from the hydrolyzate of starch without the addition of organic solvents34.
Cellulases
Commercially available cellulases display optimum activity over a pH range from 4 to 6. No enzyme with an alkaline pH optimum for activity (pH 10 or higher) had been reported before our finding of alkaline cellulase. The found alkaliphilic bacteria (Bacillus sp. No. N-4 and No. 1139) producing extracellular carboxymethylcellulases (CMCases) 35, 36. The alkaliphilic Bacillus sp. No. N-4 (ATCC21833), produces multi-CMCases which are active over a broad pH range (pH 5 to 10). Two alkaline CMCases (enzymes E1 and E2) with an optimum pH for enzyme action at pH 10.0 have been partially purified from the crude enzyme preparation. The enzyme E2 is stable up to 80°C and E1 up to 60°C. No differences between the two enzymes have been observed so far in the type of products formed. Sashihara37 cloned the cellulase genes of Bacillus sp. No. N-4 in Escherichia coli HB101 with pBR322.
Beppu and coworkers (REF) constructed many chimeric cellulases from B. subtilis and Bacillus sp. strain N-4 enzyme genes in an effort to understand the alkaliphily of N-4 enzymes. Although the genes have high homology, the pH activity profiles of the two enzymes are quite different; the B. subtilis enzyme (BSC) has its optimum pH at 6 to 6.5, whereas the Bacillus sp. Strain N-4 enzyme (NK1) is active over a broad pH range from 6 to 10.5. The chimeric cellulases showed various chromatographic behaviors, reflecting the origins of their C-terminal regions.
Enzymes are also increasingly playing a key role in the finishing of fabrics and clothes. In the technique popularly called biopolishing, cellulases eliminate the rough cellulose lumps formed on cloth, thereby providing an even finish to the fabric as well as a brighter color. A similar finishing effect is produced when the enzyme is included as a laundry detergent additive. Traditionally, stonewashing of denims had been carried out using pumice stones, which damage cloth. In the present technology, cellulases have replaced pumice stones, resulting in the non-abrasive process known as biostoning. The biostoning of denim generally involves the use of acid and neutral cellulases, which have the undesirable property of causing back staining of indigo dye. Alkaline or alkali-stable cellulases, which can diminish indigo back staining under higher pH, are recommended for further improvement of the process38.
Lipase
Lipases cause hydrolysis of triglycerides releasing fatty acids and glycerol. They can also catalyze the reverse esterification reactions, thus producing glycerides from glycerol and fatty acids. Many lipases are also involved in the catalysis of trans-esterification reactions and enantioselective hydrolyses39,40,41,42. Most commercial lipases are obtained from fungi (mainly Rhizopus, Candida, and Rhizomucor) and bacteria (Pseudomonas and Chromobacterium)43. The optimum pH of the two lipases was 9.5. Both enzymes were inhibited by bile salts such as sodium cholate, sodium deoxycholate, and sodium taurocholate at 0.25%.Although the initial motivation for studying alkaline lipase was its application to detergents, many alkaline lipases are significantly inhibited in the presence of either alkylbenzene sulfate or dodecyl benzene sulfonate3. Watanabe44 conducted an extensive screening for alkaline lipase-producing microorganisms from soil and water samples. Two bacterial strains were selected as potent producers of alkaline lipase. These were identified as Pseudomonas nitroreducens nov. subsp. thermotolerans and P. fragi. Wang45 then produced thermostable alkaline lipase from a newly isolated thermophilic Bacillus, strain A30-1 (ATCC 53841). The organism grew optimally at 60-65°C and in the pH range of 6-9. It was characterized as a Bacillus species.
Xylanase
The first paper of xylanase production by an alkaliphilic bacteria was published in 1973 by Horikoshi and Atsukawa46. Okazaki reported that four thermophilic alkaliphilic Bacillus stains (Wl (JCM2888), W2 (JCM2889), W3 and W4) produced xylanases47 . The pH optima for enzyme action of strains Wl and W3 was 6.0 and for strains W2 and W4 was found to be between 6 and 7. The enzymes are stable between pH 4.5 and 10.5 at 45°C for 1 h. The optimum temperatures of xylanases of Wl and W3 is 65°C and those of W2 and W4, 70°C. The degree of hydrolysis of xylan is about 70% after 24 h incubation. The purified enzyme of Bacillus sp. No. C- 59-2 exhibits a broad optimum pH ranging from 6.0 to 8.0. Xylanases are hydrolases with a substantial market value of US $200 million48. Since xylan contains many side chains, several enzymes are required to act in a synergistic fashion to effect complete hydrolysis of the substrate.
Pectinase
Pectinolytic enzymes (pectinases), which degrade pectic polysaccharides such as pectin and pectic acid, are distributed in microorganisms and higher plants but not higher animals. Microbial pectinases are widely used in the fruit- and vegetable-processing industries. Recently, a novel field of application is envisaged for pectinases in the production of oligosaccharides as functional food components49. Fogarty and coworkers50, 33 have also reported that Bacillus sp. strain RK9 produces an endopolygalacturonate lyase, whose optimum pH for the enzyme activity toward acid-soluble pectic acid was 10.0.
Bacteria secreting alkaline pectinase were first used in the retting process of Mitsumata bast52. Alkaliphilic bacterial strain NT-33 can also degum ramie fibers53 and the retting process of a type of Japanese paper has been improved considerably by the use of alkaline pecticlyase (optimal pH of 9.5) from Bacillus sp. strain GIR 277; this use leads to the production of stronger, better quality paper. Polysaccharide-degrading enzymes from alkaliphilic bacteria have been studied for their effectiveness in degumming of ramie fibers54 and thermoalkali-stable polygalacturonase from Bacillus sp. MG-cp-2 are used for similarly treating ramie and sunn hemp fibers55 .
Catalase
Catalases are classified into: monofunctional catalases, catalase-peroxidases with dual functionality, non-heme catalases, and minor catalases. In addition there exist minor catalases, such as chloroperoxidase and plant peroxidases, are heme-containing proteins. Monofunctional catalases comprise the largest group and are produced by most aerobic Prokaryotes and Eukaryotes. Structurally, they are homotetramers with four prosthetic heme groups and range in size from 200 to 340 kDa. The catalase-peroxidases are homodimeric heme-containing proteins of sizes 120–340 kDa and display both catalase and peroxidase activities. Peroxidase (EC 1.11.1.7) function results in the reduction of H2O2 to H2O utilizing organic substances as electron donors56. Non-heme catalases contain manganese (and not heme) in their active sites and have been isolated from diverse groups of bacteria; catalases find biotechnological applications in the food, medical, and textile industries57.
Catalase (EC 1.11.1.6) is an anti-oxidant biocatalyst facilitating the conversion of H2O2 to O2 and H2O58, and is produced in response to oxidative stress or due to presence of ROS59. Aerobic organisms utilize oxygen to facilitate efficient metabolism of nutrients. However, during this process, oxidants (collectively called reactive oxygen species or ROS) are formed. The ROS are formed due to incomplete reduction of oxygen60. Such oxidants can cause damage to lipids, proteins, and nucleic acids, as a result, microorganisms produce antioxidants in the form of enzymes and other molecules to reduce the levels of ROS60..
Allgood and Perry61 in 1986 reported that thermo- and alkali-stable catalases such as that produced by Thermoleophilum album. Yumoto’s group, in 1990, had reported of alkaliphilic Bacillus YN-2000 secreting a catalase that showed substantial peroxidase as well as catalase activity. The catalase quantity was found to be elevated when cells were grown at pH 10.0 than at lower pH ranges of 7.0–9.0. Hicks62 observed that cells of B. firmus OF4, a facultative alkaliphile, showed twice the specific activity for catalase when grown at pH 10.5 compared to when grown at pH 7.5. Nevertheless, the cells grown at pH 10.5 showed more sensitivity to exogenous hydrogen peroxide. In the facultative psychrophile Vibrio rumoiensis S-1T, isolated from an H2O2-rich location63, the catalase activity was found to be 527,500 U mg protein-1, and was faster than that of bovine liver catalase. The catalase functioned optimally at broad pH ranges (6.0–10.0). Subsequently, Phucharoen57 isolated an alkali- and halo-tolerant bacterium Halomonas sp. SK1, which produced a catalase with high specific activity (57,900 U mg protein-1). This activity was twice as high as bovine liver catalase and the enzyme was active over pH ranges of 5.0–11.0 with optimal activity at pH 10.0. They concluded that use of the catalase had a noticeable and enhanced effect on the dyeing process in addition to improved color yield. There was also added simplicity of application since the catalase could be used in the dyeing bath right after bleaching, leading to lesser washing, water consumption, and effluent volume. The varied properties of catalase, notably the thermoalkali stable types, means that these enzymes are likely to continue to play an important role in textile and food-processing industries; genetic and protein engineering techniques are also likely to bring about desirable changes in the properties of catalases, making them even more useful for use in industry.
Chitinase
Chitinous waste (chiefly from the seafood industry) needs to be recycled to maintain the carbon–nitrogen balance in the environment64. Tsujibo65 isolated two types of chitinases from the alkaliphilic Nocardiopsis albus subsp. prasina OPC-131. The optimum pH of chitinase A was pH 5.0, and that of chitinase B was pH 7.0. Later, Bhushan and Hoondal66 isolated the alkaliphilic, chitinase-producing Bacillus sp. strain BG-11. The purified chitinase from this organism exhibited a broad pH and temperature optima of 7.5 to 9.0 and 45 to 55°C, respectively, and the enzyme was found to be stable between pH 6.0 and 9.0 at 50°C for more than 2 h. Ag1, Hg21, dithiothreitol, β-mercaptoethanol, glutathione, iodoacetic acid, and iodoacetamide inhibit the activity of the enzyme up to 50%. Chitinase characteristics of gammaproteobacteria (obtained from an alkaline, hypersaline lake) and from a metagenomic library (estuarine bacteria) have been studied using fosmids, the latter are f-factor cosmids capable of containing up to 50-kb DNA. The enzymes from the alkaline lake organisms showed distinctive adaptations making them haloalkali tolerant.
References
- Horikoshi, K. General view of alkaliphiles and thermophiles. In Superbugs: Micro–organisms in Extreme Environments. (Eds. K. Horikoshi and W.D. Grant), Springer Verlag. Berlin. 1991; 3-13.
- Sarethy, I.P., Y. Saxena, A. Kapoor, M. Sharma, S.K. Sharma, V. Gupta, and S. Gupta. Alkaliphilic bacteria: applications in industrial biotechnology. J. Ind. Microbiol. Biotechnol. 2011; 38:769–790.
- Horikoshi, K. Alkaliphiles: some applications of their products for biotechnology. microbiology and molecular biology reviews. 1999; 63: 735-50.
- Horikoshi, K. Production of alkaline enzymes by alkalophilic microorganisms. Part 1. Alkaline protease produced by Bacillus No, 221. Biol. Chem. 1971; 36: 1407- 1414.
- Krulwich, T.A and A.A. Guffanti. Alkalophilic bacteria. Ann. Rev. Microbiol. 1989; 43: 435-463.
- Duckworth, A.W., W.D. Grant, B.E. Jones and R. Vansteenbergen. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol. Ecol. 1996; 19:181–191
- Kanekar, P.P., S.S. Nilegaonkar, S.S. Sarnaik, and A.S. Kelkar. Optimization of protease activity of alkaliphilic bacteria isolated from an alkaline lake in India. Bioresource technology. 2002; 85: 87-93.
- Van den Burg, B. Extremophiles as a source for novel enzymes. Curr.Opin.Microbiol. 2003; 6: 213–218.
- Grant, W.D., Tindall, B.J. The isolation of alkalophilic bacteria. In Microbial Growth and Survival in Extremes of Environment, (Eds. G.W. Gould and J.G.L. Corry), 1980; pp 27-36. Academic Press. London.
- Jones, B.E., Grant, W.D., Collins, N.C., Mwatha, W.E. Alkaliphiles: Diversity and Identification. In Bacterial Diversity and Systematics, (Ed. F.G. Priest), 1994; pp 195-230. Plenum Press. New York.
- Takami, H., A. Inoue, F. Fuji and K. Horikoshi. Microbial flora in the deepest sea mud of the Mariana Trench. FEMS Microbiol. Lett. 1997;152: 279–285
- Wiegel, J. Anaerobic alkali thermophiles, a novel group of extremophiles. Extremophiles 1998; 2:257–267.
- Niimura, Y., F. Yanagida, T. Uchimura, N. Ohara, K. Suzuki and M. Kozaki. A new facultative anaerobic xylan-using alkalophile lacking cytochrome, quinone and catalase. Agric. Biol. Chem. 1987; 51: 2271–2275.
- McGenity, T.J. and W.D. Grant. The haloalkaliphilic archaeon (archaebacterium) Natronococcus occultus represents a distinct lineage within the Halobacteriales, most closely related to the other haloalkaliphilic lineage (Natronobacterium). Syst. Appl. Microbiol. 1993;16: 239–243.
- Kamekura, M., M.L. DyallSmith, V. Upasani, A. Ventosa and M. Kates. Diversity of alkaliphilic halobacteria: proposals for transfer of Natronobacterium vacuolatum, Natronobacterium magadii, and Natronobacterium pharaonis to Halorubrum, Natrialba, and Natronomonas nov., respectively, as Halorubrum vacuolatum comb. nov., Natrialba magadii comb. nov., and Natronomonas pharaonis comb. nov., respectively. Int. J. Syst. Bacteriol. 1997; 47:853–857.
- Xu, Y., P.J. Zhou and X.Y. Tian. Characterization of two novel haloalkaliphilic archaea Natronorubrum bangense gen. nov., sp. nov., and Natronorubrum tibetense nov., sp. nov. Int. J. Syst. Bacteriol. 1999; 49: 261– 266.
- Desmarais, D., P. E. Jablonski, N. S. Fedarko, and M. F. Roberts. 2-Sulfotrehalose, a novel osmolyte in haloalkaliphilic archaea. J. Bacteriol. 1997;179:3146–3153.
- Horikoshi, K. Micro-organisms in Alkaline Environments. VCH Verlagsgesellschaft mbH. Weinheim. 1992.
- Rao, M.B., A.M. Tanksale, M.S. Ghatge and V.V. Deshpande. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 1998; 62:597–635.
- Schallmey, M., A. Singh and O.P. Ward OP. Developments in the use of Bacillus species for industrial production. J. Microbiol. 2004; 50: 1–17.
- Takami, H., T. Akiba and K. Horikoshi. Production of extremely thermostable alkaline protease from Bacillus no. AH-101. Appl. Microbiol. Biotechnol. 1989; 30: 120–124.
- Y.T., J.O. Kim, S.Y. Moon, H.H. Lee and H.M. Rho. Extracellular alkaline proteases from alka- Iophilic Vibrio metschnikot’ii strain HH53. Biotechnol. Lett. 1994; 6: 413-418.
- Han, X.Q. and S. Damodaran. Purification and characterization of protease Q: A detergent- and urea-stable serine endopeptidase from Bacillus pumilus. J. Agric. Food Chem. 1998; 46: 3596–3603.
- Verma, A., M.W. Ansari, M.S. Anwar, R. Agrawal and S. Agrawal. Alkaline protease from Thermoactinomyces RS1 mitigates industrial pollution. Protoplasma. 2014; 251: 711-718.
- Ito, S., T. Kobayashi, K. Ara, K, Ozaki, S. Kawai and Y. Hatada. Alkaline detergent enzymes from alkaliphiles: enzymatic properties, genetics, and structures. Extremophiles, 1998; 2: 185–190.
- Gupta, R., Q.K. Beg and P. Lorenz. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 2002; 59:15–32.
- Anwar, A and M. Saleemuddin. Alkaline proteases: a review. Bioresour Technol. 1998; 64:175–183.
- Kumar, C.G. and H. Takagi. Microbial alkaline proteases: from a bioindustrial viewpoint. Biotechnol. Adv. 1999; 17: 561–594.
- Jaouadi, N.Z., B. Jaouadi, H.B. Hlima, H. Rekik, M. Belhoul, M. Hmidi and S. Bejar. Probing the crucial role of Leu31 and Thr33 of the Bacillus pumilus CBS alkaline protease in substrate recognition and enzymatic depilation of animal hide. PloS One. 9: e108367. 2014.
- Yamamoto, M., Y. Tanaka and K. Horikoshi. Alkaline amylases of alkalophilic bacteria. Biol. Chem. 1972; 36: 1819-1823.
- Kim, T.U., B.G. Gu, J.Y. Jeong, S.M. Byun and Y.C. Shin. Purification and characterization of a maltotetraose-forming alkaline a-amylase from an alkalophilic Bacillus strain, GM8901. Appl. Environ. Microbiol. 1995; 61: 3105–3112.
- McTigue, M. A., C. T. Kelly, E. M. Doyle and W. M. Fogarty. The alkaline amylase of the alkalophilic Bacillus sp IMD 370. Enzyme Microb. Technol. 1995; 17: 570–573.
- Kelly, C.T and W.M. Fogarty. Production and properties of polygalacturonate lyase by alkalophilic microorganisms, Bacillus RK9. Can. J. Microbiol. 1978; 24: 1164–1172.
- Matsuzawa, M., M. Kawano, N. Nakamura and K. Horikoshi. An improved method for the production of schardinger/β-dextrin on an industrial scale by cyclodextrin glycosyltransferase of an alkalophilic Bacillus Die Staerke. 1975; 27: 410-413.
- Horikoshi, K., M. Nakao, Y. Kurono and N. Sashihara. Cellulases of and alkalophilic Bacillus strain isolated from soil. Can. J. Microbiol. 1984; 30: 774-779.
- Fukumori, F., T. Kudo and K. Horikoshi. Purification and properties of a cellulase from alkalophilic Bacillus No. 1139. J. Gen. Microbiol. 1985; 131: 3339-3345.
- Sashihara, N., T. Kudo and K. Horikoshi. Molecular cloning and expression of cellulase genes of alkalophilic Bacillus strain N-4 in Escherichia coli. J. Bacteriol. 1984; 158: 503-506.
- Anish, R., M.S. Rahman and M. Rao. Application of cellulases from an alkalothermophilic Thermomonospora sp. in biopolishing of denims. Biotechnol. Bioeng. 2007; 96:48–56.
- Nini, L., L. Sarda, L.C. Comeau, E. Boitard, J.P. Dube`s and H. Chahinian. Lipase-catalysed hydrolysis of short-chain substrates in solution and in emulsion: a kinetic study. Biochim. Biophys. Acta. 2001; 1534: 34–44.
- Nagayama, K., N. Yamasaki and M. Imai. Fatty acid esterification catalyzed by Candida rugosa lipase in lecithin microemulsion- based organogels. Biochem. Eng. J. 2002; 12: 231–236.
- Piao, J., T. Kobayashi, S. Adachi, K. Nakanishi and R. Matsuno. Synthesis of mono- and dioleoyl erythritols through immobilized- lipase-catalyzed condensation of erythritol and oleic acid in acetone. Biochem. Eng. J. 2003; 14: 79–84.
- Raku, T., M. Kitagawa, H. Shimakawa and Y. Tokiwa. Enzymatic synthesis of trehalose esters having lipophilicity. J. Biotechnol. 2003; 100: 203–208.
- Dheeman, D.S., J.M. Frias and G.T.M. Henehan. Influence of cultivation conditions on the production of a thermostable extracellular lipase from Amycolatopsis mediterranei DSM 43304. J. Ind. Microbiol. Biot. 2010; 37(1): 1–17.
- Watanabe, N., Y. Ota, Y. Minoda and K. Yamada. Isolation and identification of alkaline lipase-producing microorganisms, culture conditions and some properties of crude enzymes. Agric. Biol. Chem. 1977; 41:1353– 1358.
- Wang, Y.X., K.C. Srivastava, G.J. Shen and H.Y. Wang. Thermostable alkahne lipase from a newly isolated thermophilic Bacillus, strain A30-1 (ATCC 53841). J. Ferment. Bioeng. 1995; 79: 433-438.
- Horikoshi, K and Y. Atsukawa. Xylanase produced by alkalophilic BacillusC- 59-2. Agr. Biol. Chem. 1973; 37: 2097-2103.
- Okazaki, W., T. Akiba, K. Horikoshi and R. Akahoshi. Production and properties of two types of xylanases from alkalophilic thermophilic Bacillus Appl. Microb. Biotechnol. 1984; 19: 335-340.
- Katapodis, P., V. Christakopoulou, D. Kekos and P. Christakopoulos. Optimization of xylanase production by Chaetomium thermophilium in wheat straws using response surface methodology. Biochem. Eng. J. 2007; 35: 136–141.
- Horikoshi, K. 2006. Alkaliphilies: genetic properties and application of enzymes.
- Fogarty, W.M and P.O. Ward. Pectinases and Pectic Polysaccharides, 13. Churchill Livingstone, Edinburgh, United Kingdom1977.
- Yoshihara, K. and Y. Kobayashi. Retting of Mitsumata bast by alkalophilic Bacillus in paper making. Agric. Biol. Chem. 1982; 46: 109–117.
- Cao, J., L. Zheng and S. Chen. Screening of pectinase producer from alkalophilic bacteria and study on its potential application in degumming of ramie. Enzyme. Microb. Technol. 1992; 14: 1013–1016.
- Zheng, L., Y. Du and J. Zhang. Degumming of ramie fibres by alkalophilic bacteria and their polysaccharide-degrading enzymes. Biores. Technol. 2001; 78: 89–94.
- Kapoor, M., Q.K. Beg, B. Bhushan, K. Singh, K.S. Dadhich and G.S. Hoondal. Application of an alkaline and thermostable polygalacturonase from Bacillus MG-cp-2 in degumming of ramie (Boehmeria nivea) and sunn hemp (Crotalaria juncea) bast fibres. Process. Biochem. 2001; 36: 803–807.
- Gudelj, M., G.O. Fruhwirth, A. Paar, F. Lottspeich, K.H. Robra, A. Cavaco-Paulo and G.M. Gubitz. A catalase-peroxidase from a newly isolated thermoalkaliphilic Bacillus with potential for the treatment of textile bleaching effluents. Extremophiles. 2001; 5: 423–429.
- Phucharoen, K., K. Hoshino, Y. Takenaka and T. Shinozawa. Purification, characterization, and gene sequencing of a catalase from an alkali- and halo-tolerant bacterium, Halomonas SK1. Biosci. Biotechnol. Biochem. 2002; 66: 955–962.
- Brioukhanov, A.L., A.I. Netrusov AI, R.I. Eggen. The catalase and superoxide dismutase genes are transcriptionally up-regulated upon oxidative stress in the strictly anaerobic archaeon Methanosarcina barkeri. Microbiology. . 2006; 152: 1671–1677.
- Chelikani, P., I. Fita and P.C. Loewen. Diversity of structures and properties among catalases. Cell. Mol. Life. Sci. 2004. 61: 192–208.
- Turrensa, J.F. Superoxide dismutase and catalase. Compr. Toxicol. . 2010; 4: 219–227.
- All good, G.S. and J.J. Perry. Characterization of a manganese containing catalase from the obligate thermophile Thermoleophilum album. J. Bacteriol. 1986;168: 563–567.
- Hicks, D.B. Purification of three catalase isozymes from facultatively alkaliphilic Bacillus firmus Biochim. Biophys. Acta. 1995;1229: 347–355.
- Yumoto, I., D. Ichihashi, H. Iwata, A. Istokovics, N. Ichise, H. Matsuyama, H. Okuyama, K. Kawasaki. Purification and characterization of a catalase from the facultatively psychrophilic bacterium Vibrio rumoiensis S-1(T) exhibiting high catalase activity. J. Bacteriol. 2000; 182: 1903–1909.
- Nawani, N.N and B.P. Kapadnis. Chitin degrading potential of bacteria from extreme and moderate environment. Indian. J. Exp. Biol. 2003; 41: 248–254.
- Tsujibo, H., Y. Yoshida, K. Miyamoto, T. Hasegawa and Y. Inamori. Purification and properties of two types of chitinases produced by an alkalophilic actinomycete. Biosci. Biotechnol. Biochem. 1992; 56: 1304–1305.
- Bhushan, B and G.S. Hoondal. Isolation, purification and properties of a thermostable chitinase from an alkalophilic Bacillus BG-11. Biotechnol. Lett. 1998; 20: 157–159.
- LeCleir, G.R., A. Buchan, J. Maurer, M.A. Moran and J.T. Hollibaugh. Comparison of chitinolytic enzymes from an alkaline, hypersaline lake and an estuary. Environ. Microbiol. 2007; 9: 197–205.

This work is licensed under a Creative Commons Attribution 4.0 International License.





