How to Cite | Publication History | PlumX Article Matrix
Genetic Variations in Two Casen Genes Aamong Maghrabicamels Reared in Egypt
Othman E. Othman1*, Amira M. Nowier2 and Medhat E. El-Denary3
1Cell Biology Department, National Research Centre, Dokki, Egypt 2 Biotechnology Research Department, Animal Production Research Institute, Dokki, Egypt 3 Genetic Department, Agriculture Faculty, Tanta University, Tanta, Egypt. Corresponding Author Email : othmanmah@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2057
ABSTRACT: Camels play an important socio-economic role within the pastoral and agricultural system in the dry and semidry zones of Asia and African where they are dual purpose animals (meat and milk).In spite of the effective role of casein genes with their polymorphisms on quantitative traits and technological properties of milk, the studies on genetic polymorphism of camel milk genes are limited. This work aimed to identify the genetic polymorphisms and SNPs of two casein genes in Maghrabi camel breed in Egypt. The amplified fragments at 488-bp of κ-CN gene were digested with AluIendonuclease. The results showed the presence of three genotypes; CC (12%), TT (48%) CT (40%). The sequence analysis of two detected alleles declared the presence of a SNP (C→T) at position 121 in amplified fragments.The nucleotide sequences of κ-CN alleles C and T were submitted to GenBank with accession numbers; KU055605 and KU055606, respectively. The primers used in this study amplified 942-bp fragments of αs1-casen gene. The results of SmlI digestion did not showed any restriction site whereas the digestion with AluI endonuclease revealed the presence of two restriction sites AG^CT at positions 68^69 and 631^632 in amplified fragments. The nucleotide sequence of monomorphic αs1-casein gene was submitted to GenBank with accession number KU145820. In conclusion, the genetic characterization of genes associated with milk yield and composition in camel is considered an essential step towards its genetic improvement through the selection of superior animals depending on the favorable alleles and genotypes; marker assisted selection (MAS).
KEYWORDS: Genetic polymorphism; SNP; Maghrabicamel; κ-casen gene; αs1-casen gene
Download this article as:| Copy the following to cite this article: Othman O. E, Nowier A. M, El-Denary M. E. Genetic variations in two casein genes among Maghrabicamels reared in Egypt. Biosci Biotech Res Asia 2016;13(1) |
| Copy the following to cite this URL: Othman O. E, Nowier A. M, El-Denary M. E. Genetic variations in two casein genes among Maghrabicamels reared in Egypt. Biosci Biotech Res Asia 2016;13(1). Available from: https://www.biotech-asia.org/?p=7584 |
Introduction
A great interest has been directed to camels in the world; the camel is a very important animal in the arid and semi-arid regions. The survival of millions of human being is dependent on the camel in such areas for meat, milk and hair production and still an important mean of drought and transportation for large sectors of pastoral societies (El-Sawalhyet al., 1996). In spite of camel’s considerable contribution to food security in semi dry and dry zones, and its being a major component of the agro-pastoral systems in vast pastoral areas in Africa and Asia, little is known about its production potential and systems compared to other domestic animals. However, most previous research conducted on camels was oriented towards diseases, reproductive physiology and characterization (Mehariet al., 2007).
In Egypt, camels areimportant animals because they are dual purpose animals (meat and milk production). In theNile Valley and Delta, they are mainly raised for meat production and someagricultural labors. In the desert, they are raised equally for meat and milkproduction, some labors and transport. On the other hands, some breeders raise them for camelracing. It was reported that many camel breeds are reared in Egypt, but the main camel breeds are Maghrabi (a dualpurpose animal), however, Falahy, Sudany and Mowaled (meat type animal)(Mahran, 2004).
Recently, genetic polymorphisms at candidate genes affecting economic traits have stimulated substantial research interest because of their impending utilizationas an aid to genetic selection and to demarcate evolutionary relationships in different livestock breeds (Sodhiet al., 2007). Association of several polymorphic sites (SNPs) in different candidate genes with economic traits has been much investigated in different animal species. Studies on characterization of candidate genes and their polymorphism association with animal performance in camels are meager compared with other livestock; cattle (Lucyet al., 1991; Schleeet al., 1994; Geet al., 2003), sheep(Wallis et al., 1998; Bastoset al., 2001)and goats (Wallis et al., 1998; Gupta et al., 2007).
The casein fraction of ruminant milk proteins consists of four caseins, namely αs1, αs2, β and κ-casein. These four caseins are the main components (76-86%) of total milk protein (Swaisgood, 1992). The relative amounts of these four casein fractions affect the physicochemical, nutritional and technological properties of ruminant milks (Ramunnoet al., 2000). The casein proteins include three main specific proteins which are the calcium-sensitive (αs1-, αs2- and β-caseins) that coalesce with κ-casein, calcium and phosphate to form micelles. These casein proteins encoded by four clustered genes in a 250-kb genomic DNA fragment; αs1 is very close to β followed by αs2 and κ-caseins (Provotet al., 1995).
Despite of the important role of casein genes and their effects on quantitative traits and technological properties of milk, few studies were focused on the genetic characterization of casein genes in camels comparing with other in ruminants. The present study aimed to identify the genetic variations (polymorphisms) in two casein genes; κ- and αs1-casein genes in Maghrabi camel breed reared in Egypt using PCR-RFLP and nucleotide sequence analysis.
Materials and Methods
Animals and genomic DNA extraction
The blood samples used in this study were collected from 50 Maghrabicamel females belonging to different farms; the camel production station in MarsaMatrouh (Animal Production Institute, 25 samples) and three private farms in West Desert of Egypt (25 samples).Genomic DNA was extracted from the whole bloodaccording to the method described by Miller et al. (1988) withminor modifications. Briefly, blood samples were mixed withcold 2x sucrose-triton and centrifuged at 5000 rpm for 15 minat 4°C. The nuclear pellet was suspended in lysis buffer,sodium dodecyl sulfate and proteinase K and incubatedovernight in a shaking water bath at 37°C. Nucleic acids wereextracted with saturated NaCl solution. The DNA was pickedup and washed in 70% ethanol. The DNA was dissolved in1x TE buffer. DNA concentration was determined, using NanoDrop1000 (Thermo Scientific Spectrophotometer) and thendiluted to the working concentration of 50 ng/μlwhich issuitable for polymerase chain reaction.
Polymerase chain reaction
A PCR cocktail consisted of 1.0 μM of upper and lowerprimer specific for each tested gene, 0.2 mMdNTPs, 10x PCR reaction buffer and1.25 units of Taqpolymerase (Fermentas). The cocktail wasaliquot into PCR tubes with 100 ng of camel DNA. Thereaction was run according to the optimum condition specific for each primer (Table 1). ThePCR products were subjected to electrophoresis on 2% agarosegel stained with ethidium bromide to test the amplificationsuccess.
Restriction fragment length polymorphism (RFLP)
The PCR products for each tested genes were digested with AluI and SmlIrestriction enzymes. Ten µl of PCR product were digested with 1µl of FastDigest restriction enzymefor 15 min at the optimum temperature for maximum activity of each restriction enzyme.Gels were visualized under UV light and documented in FX Molecular Imager apparatus (BIO-RAD). Molecular size of the digested fragmentswere measured by analyzing gel images with GelAnalyzer software package version 2010a (freeware) with 100 bp DNA ladder (Larova GmbH-Germany) as DNA size marker.
Sequence Analysis
The PCR products-representatives for each detected genotype of each tested gene – were purified and sequenced by Macrogen Incorporation (Seoul, Korea). Sequence analysis and alignment were carried out using ClustalW2 to identify each single nucleotide substitution between different detected genotypes. Results of endouclease restriction were carried out using FastPCR. The nucleotide sequence of each genotype for camel κ-casein and αs1-casein genes were submitted to GenBank (NCBI, BankIt).
Table 1: The sequences and information of primers used in this study
|
Gene |
Primer sequence
5′ ——– 3′ |
PCR
Conditions (35 cycles) |
PCR product
size |
Restriction enzyme
used |
References |
|
κ-casein |
CAC AAA GAT GAC TCT GCT ATC G
GCC CTC CAC ATA TGT CTG |
94°C 1 min
56°C 1 min 72°C 2min |
488-bp |
AluI
SmlI |
Pauciullo
et al. (2013) |
|
αs1-casein |
TGA ACC AGA CAG CAT AGA G
CTA AAC TGA ATG GGT GAA AC |
94°C 1 min
54°C 1 min 72°C 1 min |
942-bp |
SmlI
AluI |
Shuiep
et al. (2013) |
Results and Discussion
Camels have an important role as meat and milk sources for many humans in different countries. The camel populations in Somalia and Sudan constitute a half of world camel populations (Pauciulloet al., 2013). The camel population in Egypt was estimated to be 120.000 headsand its ecotypesserve numerous functions in their respective production systems (e.g. milk and meat production, racing, riding and packing) and are bred and selected for sustainable performance(Mahran, 2004).
The total protein contents of camel’s milk ranged from 2.4% to 5.3% (Konuspayevaet al., 2009; Al haj & Al Kanhal, 2011; Nikkah, 2011) and it is divided into casein and whey proteins. The casein fraction constitutes 52% to 89% of total camel milk protein and it divided into 4 fractions namely αs1, αs2, β and κ-caseins which encoded by four tightly genes (Kappeleret al., 1998).
In spite of the important role of casein genes and the effects of their genetic polymorphisms on quantitative traits and technological properties of milk, the studies for the detection of genetic polymorphism of camel milk genes are still limited. Due to this fact, this work focused – using PCR-RFLP and sequencing – on the identification of genetic polymorphisms of two casein genes in Maghrabi camel breed which is a dual purpose camel breed in Egypt.
κ-casen gene
κ-casein (κ-CN) is highly heterogeneous, soluble in the presence of calcium and differs considerably in structure from the calcium sensitive caseins (Fox &McSweeny, 2003). Kappa casein is essential for micelle formation and stabilization, so it influences the manufacturing properties of milk. Cheese making is based on the cleavage of the κ-CN Phenylalnine105-Methionine106 peptide bond by enzymes or heat (Yahyaouiet al., 2001). The κ-CNfraction constitutes 3.5% of total caseins in camel milk (El Agamy, 2006). Five different isoforms of κ-CNwere found in camel milk due to a strong glycosilation of this protein. Genetic variations at DNA level of κ-casein in Somali camels did not showed any polymorphism (Kappeleret al., 1998). Due to the rare results in this field, our study aimed to detect the genetic polymorphism in exon 1 of κ-casein gene in Maghrabi came reared in Egypt.
The primers used in this study amplified 488-bp fragments (Pauciulloet al., 2013) which spans from -137 bp of 5’-flanking region to +351 bp of the camel κ-CN gene.The amplified fragments were digested with two different restriction enzymes; SmlI and AluI. The results of SmlI digestion did not showed any differentiation between tested animals where there is no any restriction site for this endonuclease in the amplified fragments. Regarding to AluI, the results showed the appearance of three different genotypes in the tested animals; CC with four digested fragments at 203-, 127-, 120- and 38-bp, TT with three digested fragments at 203-, 158- and 127-bp and CT with five digested fragments at 203-, 158-, 127-, 120- and 38-bp.
The representative samples for each detected genotype were sequenced and the results declared the presence of a single nucleotide polymorphism (C→T) at position 121 in the amplified fragments which is responsible for the destruction of restriction site (AG/CT) at this position in allele T and resulted in the presence of two different alleles C(32%) and T (68%) (Fig. 1) with three different genotypes CC(12%), CT (40%) and CT (48%) (Fig. 2).The nucleotide sequences of κ-CN alleles C and T were submitted to GenBank with the accession numbers; KU055605 and KU055606, respectively.
Pauciulloet al. (2013)reported the same SNPT>C in exon 1 of C. dromedariesκ-CNafter the digestion of amplified fragment with AluI restriction enzyme in four Sudanese breeds. They detected three different genotypes; CC (18.09%), TT (42.55%) and CT (39.36%). This finding agrees with our results where the genotype TT has the highest frequency followed by CT genotype and finally the CC genotype with the lowest frequency.
Kappa-casein gene polymorphism and its association with milk production traits was identified in cattle (Gouda et al., 2013; Deb et al., 2014), buffalo (Otavianoet al., 2005; Othman, 2005; Abbasiet al., 2009), sheep (Yousefiet al., 2013; Othman et al., 2013a) and goat (Kiplagatet al., 2010; Jemmali, et al., 2013).
 |
Figure 1: The sequence alignment between two different alleles C and T using Clustal W2 |
 |
Figure 2: The SNP (C→T) in the three detected different genotypes |
αs1-casein gene
αs1-casein (αs1-CN) is a structural component of the casein micelle, and plays an essential role in cheese curd formation (Walstraet al., 1984). αs1-CNconstitutes the second fraction of camel milk protein after β-casein. This casein gene showed different genetic variations in ruminants depending on the presence of deletions or substitutions in the triple code of amino acids (Clement et al., 2006; Chessaet al., 2010). αs1-casein polymorphism affect the milk lipids and proteins compositions, so it has a strong impact on nutritional quality and technological properties of milk (Ollieret al.,2008).The present study examined the genetic polymorphism of αs1-casein gene in Maghrabi came reared in Egypt. The primers used in this study amplified 942-bp fragmentsspanning from exon 4 to exon 6 (Shuiepet al., 2013).
The amplified fragments were digested with two different restriction enzymes; SmlI and AluI. The results of SmlI digestion did not showed any restriction site whereas the digestion with AluI endonuclease revealed the presence of two restriction sites AG^CTat positions 68^69 and 631^632 (Figs. 3 and 4) yielding the presence of three digested fragments with sizes 68-, 563- and 293-bp.
 |
Figure 3: The nucleotide sequence of 942-bp amplified fragment of αs1-casen gene AG^CT restriction sits at positions 68^69 and 631^632 in red |
 |
Figure 4: The sequences analysis showed two restriction sites AG^CT A (68^69) and B (631^632) |
The sequence alignment of αs1-casein gene in Maghrabi camel with the published sequence (Accession No.: JF429140) declared the similarity at 99% with only one SNP (A→C) at position 125 (Figs 5 and 6)
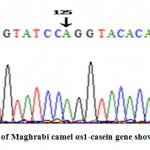 |
Figure 5: The sequence analysis of Maghrabi camel αs1-casen gene showed A nucleotide at position 125 |
Molecular characterization of αs1-casein gene was studied in Sudanese camels PCR-RFLP by Shuiepet al. (2013). They reported a SNP (G→T) characterized for the new variant CSN1S1C of this gene where this SNP destroyed the restriction site of SmlI. This finding matches with our result where the amplified fragments of Maghrabi camels did not digested with this enzyme.
The molecular characterization and the association between αs1-casein polymorphisms with milk performance were studied in ruminants like cattle (Kishore et al., 2013; Shahllaet al., 2014), buffalo (El Nahaset al., 2013; Patel et al., 2014), sheep (Othman et al., 2013b; Ceriottiet al., 2013), goat (Soareset al., 2009; Jemmaliet al., 2012).
In conclusion, the detection of genetic polymorphism and DNA sequencing of QTL genes especially milk composition is considered the best way for enhancing milk production and composition through the selection of animal with superior traits depending on molecular markers (MAS). Due to the economically important of camel in dry and semidry region in the world, further studies on genetic polymorphism of camel milk protein genes and its association with milk traits are needed in the future for genetic improvement of camel milk production.
 |
Figure 6: The sequence alignment of maghrabi alpha s1- Casen gene with the published sequence A-C substitution at position 125 in red. |
References
- Abbasi, A.,Fayazi, J.,BeigiNasiri, M.T.,Roshanfekr, H.A.,Mirzadeh, K., Sadr, A.S. Analysis of kappa casein gene polymorphism by PCR-RFLP in buffalo population in Khouzestan Province. Res. J. Biol. Sci.,2009; 4(10): 1073-2009.
- Al-haj, O.A., Kanhal, A.I. Compositional, technological and nutritional aspects of dromedary camel milk. Int. Dairy J.,2011; 20: 811-821.
- Bastos, E.,Cravador, A.,Azevedo, J., Guedes-Pinto, H. Single strand conformation polymorphism (SSCP) detection in six genes in Portuguese indigenous sheep breed “Churra da Terra Quente”. Biotechnol. Agron. Soc. Environ., 2001; 5: 7-15.
- Ceriotti, G.,Chessa, S.,Bolla, P.,Budelli, E.,Bianchi, L.,Duranti, E., Caroli, A. Single nucleotide polymorphisms in the Ovine casein genes detected by polymerase chain reaction-single strand conformation polymorphism. J. Dairy Sci.,. 2004; 87(8): 2606-2613.
- Chessa, S.,Rignanese, D.,Berbenni, M.,Ceriotti, G., Martini, M.,Pagnacco, G., Caroli, A. New genetic polymorphisms within β- and αS2-caseins. Small Rumin. Res., 2010;88: 84-88
- Clement, P.,Agboola, S., Bencini, R. A study of polymorphism in milk proteins from local and imported dairy sheep in Australia by capillary electrophoresis. LWT-Food Sci. Technol., 2006;39: 63-69.
- Deb, R., Singh, U., Kumar, S., Singh, R.,Sengar, G., Sharma, A. Genetic polymorphism and association of kappa-casein genewith milk production traits among Frieswal (HF × Sahiwal)cross breed of Indian origin. Indian J. Vet. Res., 2014; 15(4): 406-408
- El Agamy, E.I. Camel milk. In: Handbook of non-bovine mammals. Iowa, NJ, USA: Blackwell Publisher Professional,2006; pp: 297-344.
- El Nahas, S.M.,Bibars, M.A.,Taha, D.A., El-Sayyad, H.I.Detection of two CSN1S1 variants in Egyptian buffalo. J.,Genet. Eng. Biotech.,2013; 11: 75-77.
- El-Sawalhy, A.A.,Montaser, A.M., Rizk, L.G. Diagnostic and biochemicalevaluation of camel brucellosis. Vet. Med. J., 1996; 44: 323-329.
- Fox, P.F., McSweeny, P.L.H. Advanced diary chemistry, volume 1 proteins (3rd edition, part A), KlumerAcadimic, plenium publishers. New York, USA.,2003;pp: 286- 289
- Ge, W., Davis, M.E.,Hines, H.C.,Irvin, K.M., Simmen, R.C.M.Association of single nucleotide polymorphisms in the growth hormone and growth hormone receptor genes with blood serum insulin-like growth factor I concentration and growth traits in Angus cattle. J. Anim. Sci., 2003;81: 641-648.
- Gouda, E.M.,Galal, M.Kh., Abdelaziz, S.A.Genetic variants and allele frequencies of kappa casein in Egyptian cattle and buffalo using PCR-RFLP. J. Agric. Sci., 2013; 5(2): 197-203.
- Gupta, N.,Ahlawat, S.P.S., Kumar, D., Malik, G. Single nucleotide polymorphism in growth hormone gene exon-4 and exon-5 using PCR-SSCP in Black Bengal goats – A prolific meat breed of India. Meat Sci., 2007; 76: 658-665
- Jemmali, B.,Kamoun, M.,Haddar, M., Ben Gara, A.,Selmi, H.,Hammami, M.,Amraoui, M.,Rouissi, H., Boulbaba, R.Genetic polymorphism of casein alpha-S1 gene in Tunisian local goat. Biomirror.,2012; 3(11): 48-51.
- Jemmali, B.,Ben Gara, A.,Selmi, H.,Ammari, Z.,Bouheni, C.,Ben Larbi, M.,Hammami, M.,Amraoui, M.,Kamoun, M.,Rouissi, H., Rekik, B. Kappa casein gene polymorphism in local Tunisian goats.Pakistan J. Biol. Sci., 2013; 16: 2031-2035.
- Kappeler, S., Farah, Z., Puhan, Z.Sequence analysis of Camelusdromedarius milk caseins. J. Dairy Res., 1998; 65: 209-222
- Kishore, A.,Mukesh, M.,Sobti, R.C., Mishra, B.P., Sodhi, M. Variations in the regulatory region of alpha S1-casein milk protein gene among tropically adapted Indian native (BosIndicus) cattle, 2013; http://dx.doi.org/10.5402/2013/926025
- Kiplagat, S.K.,Agaba, M.,Kosgey, I.S.,Okeyo, M.,Indetie, D.,Hanotte, O., Limo, M.K.Genetic polymorphism of kappa-casein gene inindigenous Eastern Africa goat populations. Int. J. Genet. Mol. Biol., 2010;2(1): 1-5.
- Konuspayeva, G.,Faye, B., Loiseau, G.The composition of camel milk: a meta-analysis of the literature data. J. Food Comp. Anal., 2009; 22: 95-101.
- Lucy, M.H., Hauser, S.D.,Eppard, P.J.,Krivi, G.G., Collier, R.J.Genetic polymorphism within the bovine somatotropin (bST) gene detected by polymerase chain reaction and endonuclease digestion. J. Dairy Sci., 1991; 74 (Suppl. 1), 284
- Mahran, O.M.. Some studies on blood parasites in camels (Camelusdromedarius) at Shalateen city, Red Sea Governorate. Assuit Vet. Med. J., 2004; 50(102), 172-184.
- Mehari, Y.,Mekuriaw, Z., Gebru, G.Potentials of camel production in Babilie and Kebribeyahworedas of the Jijiga Zone, Somali Region, Ethiopia. Livestock Res. for Rural Develop.,2007; 19: 4-7.
- Miller, S.A., Dykes, D.D., Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res., 1988; 16: 1215.
- Nikkah, A. Equidae, camel, and yak milks as functional foods: a review. J. Nut. Food Sci., 2011; 100-111.
- Ollier, S.,Chauvet, S., Martin, P.,Chilliard, Y., Leroux, C.Goat’s αS1-casein polymorphism affects gene expression profile of lactating mammary gland. Animal, 2008; 2(4): 566-573.
- Otaviano, A.R.,Tonhati, H.,Sena, J.A.D., Munoz, M.F.C. Kappa-casein gene study with molecular markers in female buffaloes (Bubalusbubalis). Genet. Mol. Biol., 2005; 28(2): 237-241.
- Othman, E.O.The identification of kappa-casein genotyping in Egyptian river buffalo using PCR-RFLP. Arab J. Biotech.,2005; 8(2): 265-274.
- Othman, E.O., Samia, A.E.,Nagwa, A.H, Eman, R.M., Esraa, A.B.Genetic variations of b– and k–casein genes in Egyptian sheep breeds. J. Appl. Biosci., 2013a; 64:4858-4866.
- Othman E.O., Samia A.E.,Nagwa A.H.,Eman R.M., Esraa A. B.Genetic polymorphism detection of two alpha-casein genes in three Egyptian sheep breeds. J. Genet. Eng. Biotech.,2013b; 11(2): 129-134.
- Patel, A.K., Singh, M., Suryanarayana, V.V.S.Buffalo alpha S1-casein gene 5 ‘-flanking region and its interspecies comparison. J. Appl. Genet., 2014;55(1): 75-87.
- Pauciullo, A.,Shuiep, E.S.,Cosenza, G.,Ramunno, L., Erhardt, G. Molecular characterization and genetic variability at κ-casein gene (CSN3) in camels. Gene., 2013;513(1): 22-30
- Provot, C.,Persuy M.A., Mercier, J.C.Complete sequence of the ovine β-casein encoding gene and interspeciescomparison. Gene.,1995;154: 259-263.
- Ramunno, L., Cosenza, G.,Pappalardo, M.,Pastore, N., Gallo, D., Di Gregorio, P., Masina, P.Identification of the goat CSN1S1 F allele by means of PCR-RFLP method. Anim. Genet., 2000; 31: 333-346.
- Schlee, P.,Graml, R.,Rottmann, O., Pirchner, F.Influence of growth hormone genotypes on breeding values of Simmental bulls. J. Anim. Breed. Genet.,1994; 111: 253-256.
- Shahlla, N.,Ullah, O., Sheikh, R. Genetic polymorphism of milk protein variants and their association studies with milk yield in Sahiwal cattle. African J Biotech.,2014; 13(4): 555-565.
- Shuiep, E.S.,Giambra, I., El Zubeir, I.M., Erhardt, G.Biochemical and molecular characterization of polymorphisms of αs1-casein in Sudanese camel (Camelusdromedarius) milk. Int. Dairy J., 2013; 28(2): 88-93.
- Soares, M.A.M., Rodrigues, M.T.,Mognol, G.P.,Ribeiro, L.F.C.,Silva, J.L.C., Brancalhão, R.M.C.Polymorphism of alpha s1-casein gene in a dairy goat herd in the southeastern region of Brazil. R. Bras. Zootec.,2009; 38(6): 1026-1032.
- Sodhi, M.,Mukesh, M., Prakash, B., Mishra, B.P.,Sobti, R.C.,Karn Singh, P., Singh, S., Ahlawat, S.P.S.MspI allelic pattern of bovine growth hormone gene in Indian Zebu cattle (Bosindicus) breeds. Biochem. Genet.,2007; 45: 145-153.
- Swaisgood, H.E., Chemistry of the caseins. In Advanced Dairy Chemistry, 1. Proteins. (Ed. P. F. Fox). London. 1992; pp. 63-110
- Wallis, M.,Lioupis, A., Wallis, O.C.Duplicate growth hormone genes in sheep and goat. J. Mol. Endocrin., 1998; 21: 1-5.
- Walstra, P.,Jenness, R., Badings, H.T.Dairy Chemistry and Physics. Wiley, New York, 1984; pp. 467
- Yahyaoui, M.H.,Coll, A., Sanchez, A., Josep, M., Folch, J.M.Genetic polymorphism of the caprine kappa casein gene. Appl. Sci. J. Dairy Res., 2001;68: 209-216.
- Yousefi, S.,Azari, M.A.,Zerehdaran, S.,Samiee, R., Khataminejhad, R.Effect of β-lactoglobulin and κ-casein genes polymorphism on milk composition in indigenous Zel sheep. Archiv.Tierzucht., 2013; 56(21): 216 -224.

This work is licensed under a Creative Commons Attribution 4.0 International License.





