How to Cite | Publication History | PlumX Article Matrix
Shabanali Khodashenas1, Mahdi Frouzandeh Moghadam1* and Sayed Mohammad Moazzeni2
1Department of Medical Biotechnology, School of Medical Sciences, Tarbiat Modares University, Tehran, Iran.
2Department of Medical Immunology, Faculty of Medical Science, Tarbiat Modares University Tehran Iran.
Corresponding Author E-mail: foroz@modares.ac.ir
DOI : http://dx.doi.org/10.13005/bbra/2114
ABSTRACT: Nanoparticle drug delivery systems has been attempted as novel cancer treatment approaches to overcome the limitation faced in chemotherapy and surgery. Exosomes are naturally occurring nanoparticles. Exosome targeting to HER2 positive cancer cells is dependent on ligand receptor interactions. Expression of targeting moieties in chimer with LAMP-2, which is an exosomal protein, on the surface of the exosome is a rational way to target exosomes against HER2 positive cancer cells. In the present study we devised an in silico approach to design and validate the interaction of a chimer protein of LAMP-2 and G3 DARPins with HER2 molecule. Our results indicated that the designed protein is capable of interacting with HER2 in a Herseptin like orientation. The LAMP-2/ G3 DARPins chimer designed here could be a novel candidate to target exosomes towards desired HER2 positive cancer cells. The designed exosomes could be loaded with desired drugs and targeted towards the cancer cells. Given the promising results of the conducted study more empirical studies could be carried out to further evaluation of the obtained results.
KEYWORDS: exosome; LAMP-2; DARPins; in silico
Download this article as:| Copy the following to cite this article: Khodashenas S, Moghadam M. F, Moazzeni S. M. In Silico Design and Verification of a Chimer Protein to Target Exosomes Towards HER2 Positive Cancer Cells. Biosci Biotech Res Asia 2016;13(2) |
| Copy the following to cite this URL: Khodashenas S, Moghadam M. F, Moazzeni S. M. In Silico Design and Verification of a Chimer Protein to Target Exosomes Towards HER2 Positive Cancer Cells. Biosci Biotech Res Asia 2016;13(2). Available from: https://www.biotech-asia.org/?p=11224 |
Introduction
Nano particle drug delivery systems has been attempted as novel cancer treatment approaches to overcome the limitation faced in chemotherapy and surgery. Various nanoparticles have been synthesized and applied to carry drugs and genetic materials into tumor cells. Exosomes are naturally occurring nanoparticles with 40-100 nm diameter, constitutively secreted from viable cells such as dendritic cells, B cells and tumor cells (1, 2). Within the biological fluid, they communicate with surrounding cells to convert their activity or microenvironment niche. Immune modulation, promotion of angiogenesis, and tumor metastasis are among the biological activities of the exosomes, to name but a few (3). They exert their biological activities by specific delivery of biological messenger molecules like miRNA, mRNA and protein from the origin cells to the recipient tissue(4). Exosome targeting to specific cells is dependent on ligand receptor interactions (5). Therefore, providing amenable ligand receptor pairs it would be possible to expeditiously target exosmes towards desired recipients. Alvares and his coworkers applied this concept for specific delivery of siRNA into brain cells. To this end, a specific rvg against brain cell surface protein, acetyl colin receptor, was expressed as a chimer with LAMP-2b protein which is highly expressed on exosome surface (6).
Various types of human tumors like breast, ovarian and gastric cancer overexpress human epidermal growth factor receptor (HER2) which is a tyrosine kinase activated receptor that correlates with an aggressive tumor phenotype (7-10). Humanized monoclonal antibody, trastozomab, was applied against HER2 for induction of apoptosis in cancerous cells by inhibition of tyrosine kinase activity (11, 12). Elevated expression of this receptor on the cancer cell surface suggests that HER2 could serve as a receptor target in tumor drug delivery systems. Due to the limitations of the monoclonal antibody production and its application for targeted therapy (8, 13) numerous researches have been orientated towards designing synthetic peptides against HER2.
G3 DARPins is a recently designed class of ankyrin repeated protein which was selected by ribosomal display technology with high affinity against HER2 (14). This synthetic peptide applied for HER2 positive tumor cells imaging and successfully expressed as chimeric protein with lentiviral glycoprotein and bacterial membrane protein omp-A for targeted therapy (15-17).
In the present study, we explored the probability of expressing G3 DARPins as a chimer with LAMP-2b protein on the exosome surface to target HER2 positive cancer cells. In this regard, the possibility of our hypothesis was assessed employing in silico approaches.
Materials and methods
Sequence retrieval
The sequences of the Lysosome-associated membrane glycoprotein 2 (LAMP-2) along with the sequence of the receptor tyrosine -protein kinase erbB-2 (HER2) were obtained from the Uniprot knowledgebase at http://www.uniprot.org/.
Obtaining protein structure
The PDB database at www.rcsb.org/pdb/home/home.do was employed to find the 3 dimensional (3D) structures of the LAMP-2 and HER2 molecules.
Structure edition
In order to have the suitable structure for the following analyses, the structures should be edited and the unnecessary molecules like ligands should be omitted. In this regard, Protein Data Bank File Editor software (created by Jonas Lee) was fed by the 3BE1 PDB file. PDB editor was used to omit the 3 associated ligands and the dual specific bH1 Fab which is in complex with the extracellular domain of HER2.
Protein design
The protein sequences of a signal peptide, His tag, G3 DARPins, a linker and LAMP-2 were used in order to construct the final protein.
Protein modelling
All three major approaches for protein modelling were considered to predict the 3D structure of the designed protein. Protein BLAST tool at http://blast.ncbi.nlm.nih.gov/Blast.cgi was employed to search a suitable template against PDB for homology modeling. ITASSER server at http://zhanglab.ccmb.med.umich.edu/I-TASSER/ was employed to model the protein using threading method. QUARK server at http://zhanglab.ccmb.med.umich.edu/QUARK/ was used for ab initio modeling. Robetta server at http://robetta.bakerlab.org/ was employed to model the designed model using a combination of the homology modeling and threading method.
Quality assessment and refinement
The quality if built models were assessed using QMEAN (http://swissmodel.expasy.org/qmean/cgi/index.cgi) and Prosa (https://prosa.services.came.sbg.ac.at/prosa.php) servers. The model with the best quality was selected for quality refinement. The ModLoop server at https://modbase.compbio.ucsf.edu/modloop/ was used to remodel the regions of the structure
associated with high residue error. The regions of high residue error were determined according to the residue error plot of the QMEAN results. The regions spanning the 15-25 and 150-160 amino acids were modeled using this software. The all atom refinement of the selected model was done by 3Drefine server at http://sysbio.rnet.missouri.edu/3Drefine/. The refinement was done for two iterations.
Docking analyses
The possible orientation of the interaction between the designed protein and the extracellular domain of HER2 was predicted by ZDOCK (http://zdock.umassmed.edu/) and ClusPro (http://cluspro.bu.edu/login.php) servers.
Theory validation
To check the accuracy of predicted interaction orientation, the structure of the HER2 in complex with the Herceptin Fab was obtained from PDB and visually inspected to compare with the predicted interactions.
Results
Sequences and structures
The sequences of LAMP-2 and HER2 proteins were found under the accession number of P17047 and P04626 respectively. The search for the resolved 3D structure of these proteins did not lead to any structures for LAMP-2, while the extracellular domain of the HER2 protein had a 3D structure under the accession number of 3BE1. 3 associated ligands and the dual specific bH1 Fab which is in complex with the extracellular domain of HER2 were removed from the 3BE1 structure.
Protein design
The tandem sewing of the signal peptide, His tag, G3 DARPins, a linker and LAMP-2 sequences resulted in a 538 amino acids protein. The sequence of the designed protein is presented in Figure 1
 |
Figure 1: The protein sequence of the designed protein |
Protein modeling
Since the BLAST search of the PDB for suitable templates failed to find any high coverage templates, the homology modeling approach could not be used. Ab initio approach was also inapplicable due to size limitations of the QUARK server. However, ITASSER and Robetta servers proceeded in model construction and each of them resulted in 5 models.
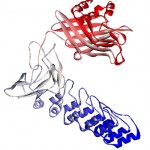 |
Figure 2: The structure of designed protein. The structure belongs to the Robetta prediction after performing the refinements. The structure is colored blue to red corresponding to the N terminus of the protein to its C terminus. |
Model assessment and refinement
The quality assessment of the best ITASSER and Robetta models indicates that the model built by Robatta server harbors the highest quality Figure 2. Therefore, it was fed as an input to the quality refinement servers. The double rounds of loop modeling and the full atomic refinements augmented the model quality. Table 1 demonstrated the quality scores of the initial and the refined models.
Table 1: The Z-scores assigned to the quality of the modeled proteins before and after refinement.
| Model | QMEAN Z-score | Prosa Z-score |
| Best ITASSER model | -5.57 | -3.48 |
| Best Robetta model | -2.74 | -7.26 |
| Refined robetta model | -2.09 | -6.95 |
Protein docking
The possible orientation of the interaction between designed protein and the HER2 extracellulr domain predicted by protein docking servers. Figure 3 illustrates that the interaction is between G3 DARPins and the HER2 protein is possible.
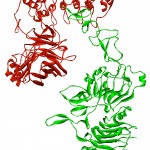 |
Figure 3: Docking result for designed protein and HER2 extracellular domain. The complex illustrates the interaction between the designed protein (red) and the extracellular domain of the HER2 (green). |
Theory validation
Figure 4 clearly demonstrate that the interacting region between the HER2 and Herceptin FAB is the same region involved in interaction with the G3 DARPins of the designed protein
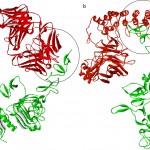 |
Figure 4: The comparison of the interaction orientation between the two complexes of HER2 extracellular domain/Herceptin fab (a) and HER2 extracellular domain/designed protein (b). The structure of the HER2 extracellular domain is in green, while Herceptin fab (a) and designed protein (b) are in red. |
Discussion
In the present study, we used LAMP-2b lysosomal associated membrane protein as an anchor for DARPins display on exosome surface. Originally this protein is expressed in exosomes and extra cellular vesicles. Initially it was used for expression of specific ligands on exosomes surface by Alvares and others. The N terminus domain of LAMP-2b is exposed on exosome surface and the ligands must be attached to this end for exosome display purposes. This could be achieved by inserting the ligand gene between the signal sequence and the mature peptide gene of the LAMP-2. Therefore, the sequences of the signal peptide, His tag, G3 DARPins, a linker and LAMP-2 sequences were sewed tandem to result in a 538 amino acids protein. The order of the sequences would make the G3 DARPins sequence to be presented on the surface of the exosome when the signal sequence is cleaved by the cellular mechanisms of protein synthesis.
Having the G3 DARPins exposed on the surface of the exosome is a biologically significant phenomena. Although chemotherapy is the most popular procedure for tumor therapy, the side effects associated with employing chemical components encouraged the researches to migrate towards packaging of therapeutics and various particles have been developed in this regard (16). Doxosome ,the doxorobicin packaged in liposome, is a successful example of packaged therapeutics used as anticancer drug, bearing low side effects (18). Other nanoparticles were also used for therapeutics packaging purposes (19) . Janson et al. are the pioneers of introducing exosomes as natural nanoparticles released from cells into biological fluids. They carry bio-messengers to recipient cells and change their functions (20). Valadi and his coworker discovered RNA entities within the exosomes and showed that they are of functional nature (21) . Based on this finding Alvarez et al. designed targeted exosomes against acetylcholine receptors on brain cells. They succeeded in transferring siRNA into brain cells and knocking down bace gene expression up to 60 percent (6). Moreover, exosomes expressing an EGFR targeting peptide
on their surfaces were employed for specific miRNA transfer into breast cancer cells (22). Exosomes were also used for specific delivery of chemical components. Tian and his coworkers used targeted exosomes for doxorubicin based therapy of breast cancer (23). Targeted exosomes could be obtained expressing specific ligands on the exosome surface which are against cell specific markers on the cell membrane. Therefore, the exosomes capable of expressing G3 DARPins on their surface could be useful to target them against plasma membranes of cancer cells expressing their receptors.
HER2 is an oncogenic tyrosine kinase growth factor receptor which is overexpressed in many metastatic and aggressive tumors. It has been reported that 20 percent of breast cancer cases overexpress HER2 (24). Thus, HER2 could be contemplated as a suitable marker in cancer detection and targeting (25, 26). Various studies have been carried out designing a amenable ligand such as antibody, nanobody, synthetic peptides like rvg and DARPins to target HER2 molecule (27-29). Herceptin, a humanized monoclonal antibody, is one of the most successful targeting agents designed for HER2 attachment. This attachment blocks tyrosine kinase pathway in order to inhibit cancer cell proliferation. DARPins (Designed Ankyrin Repeat Proteins) are a class of small proteins, designed base on natural ankyrin repeat proteins. These proteins are common binding proteins in nature and responsible for diverse functions such as cell signaling and receptor binding. Small size, high potency, high stability, high affinity (strong binding) and flexible architecture are among their advantages to name but a few. These synthetic peptides designed in 2003 and improved for HER2 targeting in 2006 by poluctum. Thereafter, they expressed in chimer with lenti and adeno virus glycoproteins for HER1 targeting on skbr3 and BT cells (30). Plückthun used this molecule for imaging in 2014, all of these experiments approved the efficiency of DARPins as ligand for targeted exosome therapy. In this regard, we
hypothesized that a G3 DARPins expressed in association with LAMP-2 protein could be a novel approach to target the exosomes against cancer cells. To assess the reliability of our hypothesis the 3D structure of the LAMP-2/ G3 DARPins were modeled and docked against the extracellular domain of the HER2 molecule employing an integrative in silico approach. In silico approaches insinuate themselves into every corner of biological research circumventing costly and time consuming experimental methods. The evaluation of this hypothesis exploiting an in silico approach is a rational choice to find out the possibility of this idea using a scientifically valid and amenable approach. Since the extracellular region of the HER2 molecule is responsible for its signaling properties, the designed protein should interact with HER2 in this region. Our analysis revealed that the designed protein could interact with HER2 molecule in a Herceptine like orientation. Since this conclusion was derived comparing the designed protein with an experimentally resolved structure of HER2/Herceptin, these results seems to be reliable. This observation could be construed as the capability of LAMP-2/ G3 DARPins chimer to target the drug containing exosomes towards HER2 positive cancer cells. Moreover, since the LAMP-2/ G3 DARPins chimer is capable of interacting with HER2 in a Herceptin like orientation, it could have the same therapeutic effects of Herceptin on the cancer cells and inhibit the proliferative properties of tumor cells. It should be tken in to consideration that the LAMP-2/ G3 DARPins could overcome many of the limitations associated with the antibody expression on exosome surface.
In conclusion it should be noted that the LAMP-2/ G3 DARPins chimer designed here could be a novel candidate to target exosomes towards desired HER2 positive cancer cells. Due to the capability of exosomes for drug delivery purposes the designed exosomes could be easily used
for cancer therapy. Given the promising results of the conducted study new horizons could be opened in targeted cancer therapy field.
Acknowledgement
The authors wish to thank TarbiatModares University for supporting the conduct of this research. The authors declare that they have no conflict of interest.
Conflict of Interest
The authors declare no conflict of interests.
Refrences
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. Journal of proteomics. 2010;73(10):1907-20.
- Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids. 2014;1841(1):108-20.
- Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. Journal of molecular medicine. 2013;91(4):431-7.
- Hannafon BN, Ding W-Q. Intercellular communication by exosome-derived microRNAs in cancer. International journal of molecular sciences. 2013;14(7):14240-69.
- Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney international. 2010;78(9):838-48.
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature biotechnology. 2011;29(4):341-5.
- Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nature Reviews Cancer. 2005;5(5):341-54.
- Nahta R, Yu D, Hung M-C, Hortobagyi GN, Esteva FJ. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nature clinical practice Oncology. 2006;3(5):269-80.
- Verri E, Guglielmini P, Puntoni M, Perdelli L, Papadia A, Lorenzi P, et al. HER2/neu oncoprotein overexpression in epithelial ovarian cancer: evaluation of its prevalence and prognostic significance. Oncology. 2005;68(2-3):154-61.
- Serrano-Olvera A, Dueñas-González A, Gallardo-Rincón D, Candelaria M, De la Garza-Salazar J. Prognostic, predictive and therapeutic implications of HER2 in invasive epithelial ovarian cancer. Cancer treatment reviews. 2006;32(3):180-90.
- Yakes FM, Chinratanalab W, Ritter CA, King W, Seelig S, Arteaga CL. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and Akt Is required for antibody-mediated effects on p27, cyclin D1, and antitumor action. Cancer research. 2002;62(14):4132-41.
- Albanell J, Codony J, Rovira A, Mellado B, Gascón P. Mechanism of action of anti-HER2 monoclonal antibodies: scientific update on trastuzumab and 2C4. New Trends in Cancer for the 21st Century: Springer; 2003. p. 253-68.
- Chames P, Van Regenmortel M, Weiss E, Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. British journal of pharmacology. 2009;157(2):220-33.
- Zahnd C, Kawe M, Stumpp MT, de Pasquale C, Tamaskovic R, Nagy-Davidescu G, et al. Efficient tumor targeting with high-affinity designed ankyrin repeat proteins: effects of affinity and molecular size. Cancer research. 2010;70(4):1595-605.
- Mironova K, Chernykh O, Ryabova A, Stremovskiy O, Proshkina G, Deyev S. Highly specific hybrid protein DARPin-mCherry for fluorescent visualization of cells overexpressing tumor marker HER2/neu. Biochemistry (Moscow). 2014;79(12):1391-6.
- Brannon-Peppas L, Blanchette JO. Nanoparticle and targeted systems for cancer therapy. Advanced drug delivery reviews. 2004;56(11):1649-59.
- Münch RC, Mühlebach MD, Schaser T, Kneissl S, Jost C, Plückthun A, et al. DARPins: an efficient targeting domain for lentiviral vectors. Molecular Therapy. 2011;19(4):686-93.
- Allen TM, Cullis PR. Liposomal drug delivery systems: from concept to clinical applications. Advanced drug delivery reviews. 2013;65(1):36-48.
- Cho K, Wang X, Nie S, Shin DM. Therapeutic nanoparticles for drug delivery in cancer. Clinical cancer research. 2008;14(5):1310-6.
- Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochimica et Biophysica Acta (BBA)-General Subjects. 2012;1820(7):940-8.
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9(6):654-9.
- Ohno S-i, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Molecular Therapy. 2013;21(1):185-91.
- Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383-90.
- Burstein HJ. The distinctive nature of HER2-positive breast cancers. New England Journal of Medicine. 2005;353(16):1652-4.
- Yang J, Lee CH, Ko HJ, Suh JS, Yoon HG, Lee K, et al. Multifunctional magneto‐polymeric nanohybrids for targeted detection and synergistic therapeutic effects on breast cancer. Angewandte Chemie International Edition. 2007;46(46):8836-9.
- Orlova A, Magnusson M, Eriksson TL, Nilsson M, Larsson B, Höidén-Guthenberg I, et al. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer research. 2006;66(8):4339-48.
- Pruszynski M, Koumarianou E, Vaidyanathan G, Revets H, Devoogdt N, Lahoutte T, et al. Targeting breast carcinoma with radioiodinated anti-HER2 Nanobody. Nuclear medicine and biology. 2013;40(1):52-9.
- Winkler J. Nanomedicines based on recombinant fusion proteins for targeting therapeutic siRNA oligonucleotides. Therapeutic delivery. 2011;2(7):891-905.
- Jamnani FR, Rahbarizadeh F, Shokrgozar MA, Ahmadvand D, Mahboudi F, Sharifzadeh Z. Targeting high affinity and epitope-distinct oligoclonal nanobodies to HER2 over-expressing tumor cells. Experimental cell research. 2012;318(10):1112-24.
- Zahnd C, Pecorari F, Straumann N, Wyler E, Plückthun A. Selection and characterization of Her2 binding-designed ankyrin repeat proteins. Journal of Biological Chemistry. 2006;281(46):35167-75.

This work is licensed under a Creative Commons Attribution 4.0 International License.





