How to Cite | Publication History | PlumX Article Matrix
B. V. Suresh Babu 1,2, P.Gunasekaran 1 ,P.Venkataraman 2, ,S. Mohana 1, R. Kiruba 1, K. Ruban 1, S. Magesh 1, CP. Indhumathi 1,C. P. Anupama1, A. K. Sheriff 1, Kavita Arunagiri 1 and K. Kaveri 1*
1Department of Virology, King Institute of Preventive Medicine and Research, Guindy, Chennai, Tamilnadu, India.
2Department of Medical research, SRM Medical college hospital and Research Centre, SRM University, Chennai, Tamilnadu, India.
Corresponding Author E-mail: kinginstitutevirology@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2132
ABSTRACT: The objective of the study was to determine the prevalence and Genotyping of RSV A and B in Chennai, Tamilnadu, south India. Human respiratory syncytial virus (RSV) is one of the most important respiratory viruses causing acute respiratory tract infections amongst children. Based on genotyping of the attachment glycoprotein (G) gene, it is divided into two groups, RSV-A and RSV-B. We tested 850 samples of patients with influenza like illness (ILI) severe respiratory illness (SARI) during the period 2011-2014 for RSV using a conventional RT-PCR and found 124 (14.5%) samples to be positive for RSV. Further sub typing by nested PCR in which 84(10%) were RSV A, 40 (4.5%) were RSV B, suggest that RSV-A was the predominant group circulating in south India during the study period. Among patients with RSV infection, 47.5% were less than 1 year age group, 29.8 % were between 1-5 year age group, 14.5% were between 6 to 14 years age group and 8 % were above 14 years age. Phylogenetic analysis all RSV-A sequences belonged to ON1 within NA1 genotype RSV-B sequences belonged to the genotype BA9 and BA12.
KEYWORDS: Respiratory Syncytial Virus; RSV infections in Chennai; RT-PCR for RSV;RSV Genotyping in India
Download this article as:| Copy the following to cite this article: Babu B. V. S, Gunasekaran P, Venkataraman P, Mohana S, Kiruba R, Ruban K, Magesh S, Indhumathi C. P. Anupama C. P. Sheriff A. K, Arunagiri K, Kaveri K. Prevalence and Molecular characterization of Circulating Respiratory Syncytial Virus (RSV) in Chennai, South India during 2011-2014. Biosci Biotech Res Asia 2016;13(2) |
| Copy the following to cite this URL: Babu B. V. S, Gunasekaran P, Venkataraman P, Mohana S, Kiruba R, Ruban K, Magesh S, Indhumathi C. P. Anupama C. P. Sheriff A. K, Arunagiri K, Kaveri K. Prevalence and Molecular characterization of Circulating Respiratory Syncytial Virus (RSV) in Chennai, South India during 2011-2014. Biosci Biotech Res Asia 2016;13(2). Available from: https://www.biotech-asia.org/?p=13798 |
Introduction
Acute respiratory tract infection (ARI) is the leading killer among children in the world (1.9 million per year), with the number of deaths occurring in developing countries 1. One fourth (2.5 millions) of the total deaths among children less than 5 years of age occur in India and approximately 20% of these are due to ARTIs 2-4. Other than that bacterial infection, influenza virus and Human respiratory syncytial virus (HRSV) have been identified as the predominant etiological agents for lower respiratory tract infections 5-7.About 20-40 % of the children hospitalized due to RSV with ARI in India 8-10. Human respiratory syncytial virus (RSV) is the second most frequent cause of respiratory infections, and the main cause of severe cases of bronchitis and pneumonia in infants and young children 11.RSV has been considered as an important viral infection, there is no antiviral treatment and effective vaccine prophylaxis is currently available against to RSV infections12, 13.
The RSV has been classified in the Pneumovirus genus of the family Paramyxoviridae, it is an enveloped virus with negative sense single stranded RNA genome that encodes for ten proteins 22-24. Glycoprotein (G) and Fusion protein (F) are most immunogenic proteins that are expressed on the virion surface, responsible for inducing production of neutralizing antibodies 14, 18, 21-23. Both within and between the major RSV subgroups, the G protein is the most changeable viral protein with a minimal conserved ectodomain that contains 2 hypervariable regions (HVRs). The second HVR (HVR2) brings the C terminus of the protein and is commonly sequenced to examine the genetic variability of Human respiratory syncytial virus strains within a given population 25-27.
The RSV has been classified into two antigenic subgroups RSV-A and RSV-B respectively, initially on the basis of the reactivity of the virus with monoclonal antibodies directed against the attachment G protein 14-16,22,24,28-30 and currently through genetic analyses 17,19,23,26,31-33. The subtypes have been further subdivided into various genotypes. To date, 11 RSV-A (ON1, GA1–GA7, SAA1, NA1& NA2) and 17 RSV-B (GB1–GB4, SAB1-SAB3& BA1–BA10) genotypes have been identified 20.
HRSV has been recognized as an important respiratory virus, there is a dearth of information on genetic diversity of HRSV strains among children in India, with few reports from North India 34-39,52. In south India RSV molecular epidemiology is narrow. Therefore, this is the first study was to determine the prevalence and molecular epidemiology of RSV A and B in Chennai, Tamilnadu, south India.
Materials and Methods
Sampling site, collection and transport of clinical specimens
During the study period from January 2011 to December 2014, 850 Influenza virus negative clinical samples (throat/Nasal swab) are subjected to detection of other respiratory viruses like, Respiratory Syncytial Virus ,Para influenza viruses, Human Metapneumo viruses, samples are collected from out patient of Chennai Government Hospitals (Saidapet Government Hospital, Stanly Government Hospital, Periyar Nagar peripheral Government Hospital, Institute of Child Health).In the present study we are included only RSV data. Throat and nasal swab were collected following standard protocols (WHO criteria), Patient demographic details, clinical symptoms were recorded. Samples were transported to the laboratory with virus transport medium (Hanks’ balanced salt solution with 200 IU penicillin ml-1, 200 mg streptomycin ml-1and 2% BSA) The present study was approved by the Institutional Ethical Committee of the King Institute of Preventive Medicine and Research, Gundy, Chennai. The present study was carried in the Dept of Virology King Institute of Preventive Medicine and Research, Guindy, Chennai.
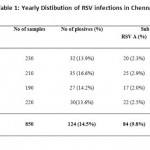 |
Table 1: Yearly Distibution of RSV infections in Chennai. |
Viral RNA Extraction
Viral RNA was isolated from the respiratory specimens using Viral RNA mini kit (Qiagen) followed by manufacture’s directions. The RNA was stored in aliquots at -80⁰C until use.
RT-PCR for detection of HRSV strains
Nucleic acid amplification was performed on GeneAmp PCR system 9600 thermal cycler (ABI- Applied Bio systems) using with Invitrogen Superscript III platinum one step RT-PCR kit for detection and sub typing of the RSV as described earlier 40. The amplified products were analyzed on a 1.5 % Agarose gel electrophoresis.
RSV Genotyping
RSV-A and RSV-B specific oligonucleotide primers (second hypervariable region of the G-protein) were used for genotyping and sequencing of the G protein. PCR protocols were described earlier 42.
RT-PCR and Nested PCR for RSV A Genotyping
Genotyping of RSV A, amplification of the second hypervariable region of the Glycoprotein gene by either external or nested PCR. The external PCR was carried out with 10 pmol of forward primer RSVA-G513-F(5′ AGTGTTCAACTTTGTACCCTGC3′) and reverse primer RSVA-F131-R (5’CTGCACTGCATGTTGATTGAT3′) 41 with Superscript III platinum one step RT-PCR kit (Invitrogen). Thermal cycling conditions are 50°C for 30 min 94°C for 5 min, followed by 40 cycles of PCR, 1 cycle consisting of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C, and a final extension is 10 min at 72°C. The 583 bp amplified products were analyzed by electrophoresis. In the case of external PCR results negative, 5 μl of the external PCR product was used as a template for nested PCR with 10 pmol of primers RSVA-G606-F (5’AACCACCACCAAGCCCACAA3′) and RSV-F22-R (5’CAACTCCATTGTTATTTGCC3′) 41, with Phusion DNA PCR kit (New England Bio labs).The cycling conditions were the same as for the external PCR (absence of RT step), except annealing temperature, which was 53°C. The nested amplicons of 391 bp were visualized by agarose gel electrophoresis.
RT-PCR for RSV-B Genotyping
For RSV B, RT-PCR assay was performed by using Superscript III platinum one step RT-PCR kit (Invitrogen) in a 25μl volume containing 20 pmol each of forward primer BGF (5’GCAGCCATAATATTCATCATCTCT3′) and reverse primer BGR (5’TGCCCCAGRTTTAATTTCGTTC3′) 41, and 5 μl of the extracted RNA. PCR reactions were conducted on a GeneAmp PCR system 9600 thermal cycler (ABI) thermal cyclic conditions are 50°C for 30 min for reverse transcription and 94°C for 5 min, 40 cycles of 94°C for 30 s, 63°C for 1 min, 72°C for 1 min, and the final extension is, at 72°C for 10 min. Amplified products were subjected to 1.5% agarose gel electrophoresis.
Sequencing and Phylogenetic Analysis
The PCR products were purified with a Purification kit (Qiagen), according to the manufacturer’s directions. The purified products were sequenced in both directions on an ABI Prism 3130 genetic analyzer (ABI) by using an ABI Prism BigDye Terminator cycle sequencing kit 3.1 (ABI). Sequences were edited manually in Bioedit software v 7.0.9.0 43. The reference global sequences were downloaded from NCBI GenBank. All the Phylogenetic analysis and multiple sequence alignment were conducted with MEGA software v 5 44.
Statistical analysis
Statistical analyses were carried out with SPSS16.0 software. Significance of differences in frequencies of age groups was tested using a chi – square test. P value was found to be statistically significant (P<0.05).
Results
Yearly Distibution of RSV infections in chennai
Out of 850 samples 124 (14.5%) samples were positive for RSV, in which 84(9.8%) were RSV A, 40 (4.7%) were RSV B, (Table I). There was an increase in the percentage positivity in 2012when compared to reaming years. During the study period from 2011-2014,
Prevalence of RSV infection in different age groups
Among patients with RSV infection, 47.5% were less than 1 year age group, 29.8 % were between 1-5 year age group, 14.5% were between 6-14 years age group and 8 % were above 14 years age (Figure I).
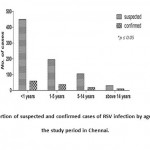 |
Figure 1: Proportion of suspected and confirmed cases of RSV infection by age group, during the study period in Chennai. |
Relation of meteorological factors with RSV infection
During the study period January 2011-December 2014 their positivity and number of samples correlated with meteorological data (Rain fall and Temperature) obtained from the Meteorological Department, Chennai, Government of India. The percentage of RSV positivity was found to be higher during the months of October to December in all the four years (Figure II).
 |
Figure 2: The correlation between meteorological variables and the number of RSV positive samples obtained during the study. |
Phylogenetic analysis
During the study period, out of 124 positive samples 27 samples (20 RSV A, 7RSV B ) are subjected to sequencing. Phylogenetic tree constructed with RSV global reference sequences and Indian sequences (Dibrugarh, Delhi and Pune, Kolkata, Lucknow ). Phylogenetic analysis detected that all RSV-A glycoprotein sequences obtained in this study clustered in ON1 group of NA1 subtypes. A newly designated genotype ON1, within the NA1 genotype was first documented from Ontario, Canada and those samples were collected in the period Dec 2010 47. Phylogenetic analysis revealed that all RSV-B glycoprotein sequences obtained in this study clustered in genotype BA, in which 4 sequences clustered in genotype BA9, 3 sequences clustered in newly designated BA12 genotype first observed in Malaysia 48 (Figure III).
 |
Figure 3: Phylogenetic Analysis of RSVA and RSV B |
Discussion
RSV is one of the most regular viral pathogens causing lower respiratory tract infections among infants, young children and elderly individuals worldwide. Different studies discovered RSV positivity in the range from 15% to 35% in pediatric population 45, 46. In India, Kolkata (Eastern India) 10.3% 39, Assam (North East) 7.9% 52, Delhi (Northern India) 17% 35, Pune (Northern India) 18.61 % 34 were detected positive, in the present study during the study period from 2011-2014 positivity rate was 14.5%. Previous studies show that RSV group A is more predominant than group B infections in globally 33, 49-51. RSV-A was the predominantly circulating group in 2011, RSV-B was the predominantly circulating in 2005–2006 while RSV-A in 2007–2008 in Kolkata 39 and RSV-A during the 2001–2005 in Delhi 35. RSV-A was the predominant circulating strain in Assam during 2009–2012 52. Our Present study shows that, 9.8% were RSV A, 4.7% were RSV B, suggesting that RSV-A was the predominant subtype circulating in Chennai, south India. Little is known regarding the seasonality of RSV in tropical and subtropical countries like India.
Most infected individuals were children less than 2 years of age. Globally, the rate of RSV infection has been reported to decrease with increasing age 46, 53, 54. We observed an increased prevalence of RSV in the less than one year (47.5%) and 1-5 years (29.8%) age groups compared with greater than 5-14 (14.5%) and above 14 years old children, which is similar to previous studies done in Kolkata, India 39. We observed that RSV infection revealed seasonal variations which peaked during the monsoon and winter months. Similar finding were reported in Pune reported a peak in monsoon and winter 34, while Kolkata and Delhi reported peaks in the winter months 36, 39. Similar findings were reported from Cambodia 46, Germany 53 and Belgium 54.
In recent years, GA2 and GA5 were the most familiar genotypes among RSV-A worldwide. GA2 was the most common genotype of RSV-A found around the world and has persisted for many years 36, 46, 53, 54. In earlier studies from Delhi, subgroups GA2 and GA5 were prevalent among the circulating RSV-A strains during the 2002 to 2003 season and genotype GA5 was predominant in the 2003 to 2005 seasons. Sequences from Pune area in 2006 clustered in genotype GA2, GA5 and the recently designated group NA2. Sequences previously documented from different parts of India clustered together in the GA2, GA3, GA5 and NA2 genotypes. Sequences from Dibrugarh, eastern India from 2009 to 2012 mainly clustered in NA1 genotype and two sequences in GA5 genotype52. In the present study not described about Amino acid analysis, this is the drawback of our study, further studies will be performed and rectify the hitch. In our study, phylogenetic analysis revealed that all RSV-A strains are clustered in newly designated ON1, within the NA1 genotype.
RSV group B genotype BA, first detected in Argentina in 1999 and has since become the predominant worldwide 36, 39, 46, 54. BA genotype is further classified from BA1 to BA12. BA genotype has been the most reported genotype in previous studies from India 35, 39. Our BA sequences clustered along with the BA9 and BA12 genotypes. Four sequences from the present study clustered in cluster BA within BA9 genotype, and three sequences in genotype BA12. However, it required long term surveillance throughout the country.
Conclusion
This is the first report contributing preliminary data on the RSV molecular characterization in Chennai, south India. In the present study concludes that RSV group A viruses predominated than group B infections during the study period in Chennai. RSV infection is pediment in the age group between less than one to five years children. In Chennai RSV infection exhibited seasonal variations which peaked during the monsoon and winter months. Phylogenetic analysis of RSV-A belonged to the ON1 within NA1 genotype, RSV-B belonged to genotype BA of BA9 and BA12. In the present study not described about Amino acid analysis, next studies will be carried out and fulfilled the hitch. However, further studies will be required from the rest of South India, which shares understanding the molecular epidemiology of RSV in south India.. RSV Genotyping data will helpful for future vaccine candidates.
Acknowledgement
The authors thank to Indian Council of Medical Research (ICMR), New Delhi for financially supported in this study. The authors acknowledge the Director, National Institute of Virology, Pune, for providing support in this influenza surveillance project and VDL staff for support in laboratory work and collection of samples.
References
- Williams, B. G., E. Gouws, C. Boschi-Pinto, J. Bryce, and C. Dye. Estimates of worldwide distribution of child deaths from acute respiratory infections. Lancet Infect. Dis., 2002;2:25–32.
- Reddaiah, V. P., and S. K. Kapoor. Acute respiratory infections in rural underfives. Indian J. Pediatr., 1988; 55:424–426.
- Ahmad, O. B., A. D. Lopez, and M. Inoue. The decline in child mortality: a reappraisal. Bull. W. H. O., 2000; 78:1175–1191.
- Rao, B. L. Epidemiology and control of influenza. Natl Med J India .,2003;16, 143–149.
- Stockton, J., Ellis, J. S., Saville, M., Clewley, J. P. & Zambon, M. C. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J Clin Microbiol.,1998; 36, 2990–2995.
- Thompson, W. W., Shay, D. K., Weintraub, E., Brammer, L., Bridges, C. B., Cox, N.J.&Fukuda, K. Influenza-associated hospitalizations in the United States. JAMA., 2004;292, 1333–1340.
- Ieven, M. Currently used nucleic acid amplification tests for the detection of viruses and atypical in acute respiratory infections. J Clin Virol., 2007; 40, 259–276.
- John, T. J., T. Cherian, M. C. Steinhoff, E. A. Simoes, and M. John. Etiology of acute respiratory infections in children in tropical southern India. Rev. Infect. Dis.,1991;S463–S469.
- Misra, P. K., R. S. Chaudhary, A. Jain, A. Pande, A. Mathur, and U. C. Chaturvedi. Viral etiology of acute respiratory infections in children in north India. J. Trop. Pediatr., 1990; 36:24–27.
- Maitreyi, R. S., S. Broor, S. K. Kabra, M. Ghosh, P. Seth, L. Dar, and A. K. Prasad. Rapid detection of respiratory viruses by centrifugation enhanced cultures from children with acute lower respiratory tract infections. J. Clin. Virol., 2000; 16:41–47.
- Borchers AT, Chang C, Gershwin ME, Gershwin LJ. Respiratory syncytial virus a comprehensive review. Clin Rev Allergy Immunol., 2013; 45: 331–379.
- Ghafoor A, Nomani NK, Ishaq Z, Zaidi SZ, Anwar F et al. Diagnoses of acute lower respiratory tract infections in children in Rawalpindi and Islamabad, Pakistan. Rev Infect Dis., 1990; 12 : S907- S914.
- McLellan JS, Yang YP, Graham BS, Kwong PD .Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat Struct Mol Biol., 2010; 17: 248–250.
- Anderson LJ, Hierholzer JC, Hendry RM, Fernie BF, Stone Y et al. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis., 1985; 151: 626–633.
- Botosso VF, Zanotto PM, Ueda M, Arruda E, Gilio AE et al. Positive selection results in frequent reversible amino acid replacements in the G protein gene of human respiratory syncytial virus. PLOS Pathog., 2009; 5:e1000254.
- Cane PA, Pringle CR Evolution of subgroup A respiratory syncytial virus: evidence for progressive accumulation of amino acid changes in the attachment protein. J Virol., 1995; 69: 2918–2925.
- Cane PA, Matthews DA, Pringle CR Analysis of respiratory syncytial virus strain variations in successive epidemics in one city. J Clin Microbiol., 1994; 32: 1–4.
- Henderson FW, Collier AM, Clyde WJ, Denny FW .Respiratory Syncytial virus infections, reinfections and immunity. A prospective, longitudinal study in young children. N Engl J Med., 1979; 300: 530–534.
- Cane PA, Matthews DA, Pringle CR .Analysis of relatedness of subgroup A respiratory syncytial virus isolated worldwide. Virus Res.,1992; 25: 15–22.
- Eshaghi A, Duvvuri VR, Lai R, Nadarajah JT, Li A et al. Genetic Variability of Human Respiratory Syncytial Virus A Strains Circulating in Ontario: A Novel Genotype with a 72 Nucleotide G Gene Duplication. PLOS ONE., 2012; 7(3): e32807.
- McLellan JS, Yang YP, Graham BS, Kwong PD .Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat Struct Mol Biol.,2010;17:248–250.
- Cane PA Molecular epidemiology of respiratory syncytial virus. Rev Med ., 2001;Virol 11: 103–116.
- Collins PL, Chanock RM, Murphy BR, DM KnipePM HowleyDE GriffinRA LambMA Martin. Respiratory syncytial virus, Vol 1, Fields virology., 2001;1443–1485.
- Goto-Sugai K, Tsukagoshi H, Mizuta KS, Matsuda M, Noda M et al. Genotyping and phylogenetic analysis of the major gains in respiratory syncytial virus isolated from infants with bronchiolitis. Jpn J Infect Dis., 2010; 63: 393–400.
- Johnson PR, Spriggs MK, Olmsted RA. Collins PL .The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci U S A ., 1987;84: 5625–5629.
- McIntosh K, Chanock RM .Respiratory syncytial virus, 1 2nd Ed, Vol 1, Fields, Virology, vol. New York, NY., 1990; pp 1045– 1072.
- Ruuskanen O, Ogra PL .Respiratory syncytial virus. Curr Probl Pediatr., 1993; 23: 50–79. [28].
- Cane PA, Pringle CR .Respiratory syncytial virus heterogeneity during an epidemic: analysis of limited nucleotide sequencing (SH gene) and restriction mapping (N gene). J Gen Virol., 1991; 72: 349–357.
- Martínez I, Valdés O, Delfraro A, Arbiza J, Russi J et al. Evolutionary pattern of the G glycoprotein of human respiratory syncytial viruses from antigenic group B: the use of alternative termination codons and lineage diversification. J Gen Virol., 1999; 80: 125– 130.
- Palomo C, Cane PA, Melero JA .Evaluation of the antibody specificities of human convalescent-phase sera against the attachment (G) protein of human respiratory syncytial virus: influence of strain variation and carbohydrate side chains. J Med Virol., 2000; 60: 468–474.
- McLellan JS, Chen M, Chang JS, Yang YP, Kim A et al. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J Virol ., 2010;84: 12236–12244.
- Melero JA, Garcia-Barreno B, Martinez I, Pringle CR, Cane PA .Antigenic structure, evolution and Immunobiology of human respiratory syncytial virus attachment (G) protein. J Gen Virol., 1997; 78(10): 2411–2418.
- Moura FEA, Borges LC, Portes SAR, Ramos EAG, Siqueira MM .Respiratory syncytial virus infections during an epidemic period in Salvador, Brazil. Viral antigenic group analysis and description of clinical and epidemiological aspects. Mem Inst Oswaldo Cruz .,2003;98: 739-743.
- Choudhary ML, Anand SP,Wadhwa BS, Chadha MS Genetic variability of human respiratory syncytial virus in Pune,Western India. Infect Genet Evol ., 2013:20: 369–377.
- Parveen S, Broor S,Kapoor SK, FowlerK, SullenderWM. Genetic diversity among respiratory syncytial viruses that have caused repeated infections in children from rural India. J Med Virol., 2006;78:659–65.
- Parveen S, Sullender WM, Fowler K, Lefkowitz EJ, Kapoor SK, Broor S. Genetic variability in the G protein gene of group A and B respiratory syncytial viruses from India. J Clin Microbiol., 2006;44:3055–64.
- Rajala MS, SullenderWM, Prasad AK, Dar L, Broor S. Genetic variability among group A and B respiratory syncytial virus isolates from a large referral hospital in New Delhi, India. J Clin Microbiol., 2003;41:2311–6.
- Yeolekar LR, Damle RG, Kamat AN, Khude MR, Simha V, Pandit AN. Respiratory viruses in acute respiratory tract infections in Western India. Indian J Pediatr., 2008; Apr;75(4):341–5.
- Agrawal, A.S., Sarkar, M., Ghosh, S., Chawla-Sarkar, M., Chakraborty, N., Basak, M., Naik, T.N.Prevalence of respiratory syncytial virus group B genotype BAIV strains among children with acute respiratory tract infection in Kolkata, Eastern India. J. ClinVirol., 2009; 45, 358–361.
- Preeti Bharaj, Wayne M Sullender, Sushil K Kabra, Kalaivani Mani, John Cherian, Vikas Tyagi, Harendra S Chahar, Samander Kaushik, Lalit Dar and Shobha Broor Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007. Virology Journal., 2009; 6:89.
- Zlateva KT, Lemey P, Moës E, Vandamme AM, Van Ranst M .Genetic variability and molecular evolution of the human respiratorysyncytial virus subgroup B attachment G protein. J Virol., 2005; 79: 9157–9167.
- Aamir UB, Alam MM, Sadia H, Zaidi SSZ, Kazi .BM Molecular Characterization of Circulating Respiratory Syncytial Virus (RSV) Genotypes in Gilgit Baltistan Province of Pakistan during 2011-2012 Winter Season. PLoS ONE., 2013; 8(9): e74018.
- Hall TA. BioEdit. A user-friendly biological sequence align ment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser., 1999; 41:95–98.
- Tamura K, Dudley J, Nei M & Kumar S .MEGA5: Molecular Evolutionary Genetics Analysis (MEGA) software version 5.0. Molecular Biology and Evolution.2007; 10.1093/molbev/msm092.
- Sato, M., Saito, R., Sakai, T., Sano, Y., Nishikawa, M., Sasaki, A., Shobugawa, Y., Gejyo,F., Suzuki, H. Molecular epidemiology of respiratory syncytial virusinfections among children with acute respiratory symptoms in a communityover three seasons. J. Clin. Microbiol.,2005; 43, 36–40.
- Arnott, A., Vong, S., Mardy, S., Chu, S., Naughtin, M., Sovann, L., Buecher, C., Beauté, J., Rith, S., Borand, L., Asgari, N., Frutos, R., Guillard, B., Touch, S., Deubel, V., Buchy, P.A study of the genetic variability of human respiratory syncytial virus (HRSV) in Cambodia reveals the existence of a new HRSV group B genotype. J. Clin. Microbiol., 2011; 49, 3504–3513.
- Eshaghi, A., Duvvuri, V.R., Lai, R., Nadarajah, J.T., Li, A., Patel, S.N., Low, D.E., Gubbay, J.B. Genetic variability of human respiratory syncytial virus a strain circulating in Ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS One., 2012; 7, e32807.
- Khor, C.S., Sam, I.C., Hooi, P.S., Chan, Y.F.Displacement of predominant respiratory syncytial virus genotypes in Malaysia between 1989 and 2011. Infect. Genet. Evol., 2013; 14, 357–360.
- Hall, C. B., E. E. Walsh, K. C. Schnabel, C. E. Long, K. M. McConnochie S. W. Hildreth, et al. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiological and clinical characteristics in hospitalized and ambulatory children. J. Infect. Dis., 1990; 162:1283– 1290.
- Gottschalk, J., R. Zbinden, L. Kaempf, and I. Heinze. Discrimination of respiratory syncytial virus subgroups A and B by reverse transcription- PCR. J. Clin. Microbiol.,1996; 34:41–43.
- Roca A, Loscertales MP, Quinto L, Perez-Brena P, Vaz N et al. Genetic variability among group A and B respiratory syncytial viruses in Mozambique: Identification of a new cluster of group B isolates. J Gen Virol., 2001; 82: 103–111.
- Dipankar Biswas, Kaushal Yadav, Biswajyoti Borkakoty, and Jagadish Mahanta. Molecular Characterization of Human Respiratory Syncytial Virus NA1 and GA5 Genotypes Detected in Assam in Northeast India, 2009–2012. Journal of Medical Virology .,2013;85:1639–1644 .
- Reiche, J., Schweiger, B. Genetic variability of group A human respiratory syncytial virus strains circulating in Germany from 1998 to 2007. J. Clin. Microbiol., 2009; 47, 1800–1810.
- Zlateva, K.T., Vijgen, L., Dekeersmaeker, N., Naranjo, C., Van Ranst, M. Subgroup prevalence and genotype circulation patterns of human respiratorysyncytial virus in Belgium during ten successive epidemic seasons. J. Clin.Microbiol., 2007;45, 3022–3030.

This work is licensed under a Creative Commons Attribution 4.0 International License.





