How to Cite | Publication History | PlumX Article Matrix
Abdollah Sadri1, Alireza Khodavandi2* and Fahimeh Alizadeh3
1Young Researchers and Elite Club, Yasooj Branch, Islamic Azad University, Yasooj, Iran.
2Department of Microbiology, Gachsaran Branch, Islamic Azad University, Gachsaran, Iran.
3Department of Microbiology, Yasooj Branch, Islamic Azad University, Yasooj, Iran.
Corresponding Author E-mail: alireza_khodavandi@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2289
ABSTRACT: Quorum-sensing quenching activityof Allium sativum, A. hirtifolium and A. cepawere screened againstCandida albicans. The morphological responseand expression patternsofselected genes involved in quorum-sensing including transcriptional repressor gene,TUP1 and hypha-specific genesHWP1, ALS1 and ALS3were examined in C. albicansatdifferent concentrations oftested extracts based on MICs. The data indicated that all extracts exerted antifungal effects through reducing the number of yeast form and inhibiting the transition from yeast to hyphae cells. Furthermore, the expression level of transcriptional repressor gene was up-regulated at different concentrations of tested extracts, which correlated with the qualitative and quantitative assessment of C. albicans. We find hypha-specific genesexhibited changes in expression at the time intervals of tested extracts exerted antifungal effectson C. albicans. These changes are in the opposite direction exhibited in transcriptional repressor gene. These are likely to be key genes in the quorum-sensing quenchingin C. albicansduring treated with tested extracts. Together, the results provide a valuable resource to quorum-sensing quenchingmechanism in C. albicans.Given the capability of A. sativum, A. hirtifolium and A. cepaextracts to effectson C. albicans and differentially expression of transcriptional repressor and hypha-specific genes, it is suggested that the tested extractsprobably have the potential to be to be used as quorum-sensing quenchers. The A. sativum, A. hirtifolium and A. cepaextracts can be usedto develop powerful new therapeutic approaches.
KEYWORDS: ALS1; ALS3; Candidaalbicans;HWP1; TUP1
Download this article as:| Copy the following to cite this article: Sadri A, Khodavandi A, Alizadeh F. Allium sativum, Allium hirtifolium and Allium cepa: The Probable Quorum-Sensing Quenching Compounds against Candida albicans. Biosci Biotech Res Asia 2016;13(3). |
| Copy the following to cite this URL: Sadri A, Khodavandi A, Alizadeh F. Allium sativum, Allium hirtifolium and Allium cepa: The Probable Quorum-Sensing Quenching Compounds against Candida albicans. Biosci Biotech Res Asia 2016;13(3). Available from: https://www.biotech-asia.org/?p=15583 |
Introduction
The polymorphic yeast Candida albicans is an opportunistic fungal pathogen found as part of the human commensal flora (Odds et al., 2003; Kruppa, 2008). Opportunistic fungal infections represent a serious threat, in an expanding population of immune- and medically compromised patients(Khodavandi et al., 2010). It is now well established that multiple virulence factors play critical role in the pathogenesisofC. albicans.These factors include transition from yeast to hyphae cells, molecules which mediate adhesion to and invasion into host cells, contact sensing and thigmotropism, biofilm formation, the ability to production of secreted hydrolytic enzymes (mainly proteinases and phospholipases), phenotypic switching (white-opaque transition) as well as evasion of host immune cells and a range of fitness attributes (Kruppa, 2008; Mayer et al., 2013; da Silva-Rocha et al., 2014).
Biofilm formation represents one of the many major virulence factors contributing to the pathogenesisof C. albicans. Biofilm formation occurs on both biological and inert surfaces through a series of steps: (i) the adhesion of yeast cells to the surface, (ii) the establishment of a microcolony along with the production of germ tubes, (iii) hyphal cells form a network with further maturation of the biofilm consisting of different cell types within an extracellular matrix, and (iv) newly formed yeast on the surface of the biofilm are released from the thick and stratified biofilm which allows further systemic dissemination of the microorganism (Kruppa, 2008; Khodavandi et al., 2011a; Nett, 2014).Many lines of evidence demonstrate that agglutinin like sequence(ALS) family and hyphal cell wall protein (HWP1) genes interfere with biofilm formation by Candida (Khodavandi et al., 2011a; Cuellar-Cruz et al., 2012).The Candida Tup1 protein is a transcription regulator that plays a role both in the regulation of the transition from yeast to hyphae cells and the transcription of a variety of virulence genes (Kebaara et al., 2008). It is relevant to highlight that the signal transduction pathways and quorum-sensing molecules play important role in biofilm development and further dissemination of infection (Kruppa, 2008; Han et al., 2011; Sharma and Jangid,2013). C. albicanswas found to produce a range of quorum-sensing molecules, including farnesol, tyrosol, farnesoic acid tryptophol, phenylethyl alcohol and morphogenic autoregulatory substance, although the precise mechanisms of action of these molecules remain unclear. Quorum-sensing has been described as a phenomenon contributing to morphogenic control in C. albicans through changes in cell density,which known as the inoculum effect. It was observed that C. albicans inoculated at cell densities below 106cells/ml under certain conditions, will germinate into the hyphal form. However, if cells densities are greater than 106 cells/ml, little germination will occur and they remain predominantly in a yeast state (Kruppa, 2008; Han et al., 2011).
Recently, therapeutic approach for quorum-sensingquenching was pioneered by quorum-sensing inhibitors. There are three ways by which the quorum-sensing signaling can be stopped/hampered: (i) stop the production of signal molecules, (ii) the signal molecules can be degraded by enzyme(s), (iii) if the signal molecule is not allowed to bind to the receptor molecule, it will not act as a regulator to the gene promoter. In either case, it would disrupt the regulation circuit. Research over the past few years has demonstrated that quorum-sensingquenching mechanisms are widely conserved in organisms and can be used for developing and formulating a new generation of antimicrobials. Many medicinal and dietary plants are known to produce quorum-sensing quenching activity. Quorum-sensing quenching activity has been reported from thegenus Allium, including Allium sativum (garlic)and A. cepa (onion) (Sharma and Jangid, 2013; Lade et al., 2014).
sativum, A. hirtifolium (shallot) and A. cepahave been used traditionally as the antimicrobial agents. It is demonstrated that A. sativum, A. hirtifolium and A. cepaextracts have been inhibitory activity against C. albicans planktonic cells and biofilms (Shuford et al., 2005; Palmeira-de-Oliveira et al., 2013; Vashisthet al., 2013; Khodavandi et al., 2014a). These results demonstrate the effects of fresh A. sativum, A. hirtifolium and A. cepaextracts as inhibitors of hyphae formation in C. albicans and suggest a possible link to quorum-sensing quenchers. The morphological responseand expression patterns ofselected genes involved in quorum-sensing including transcriptional repressor gene, TUP1 and hypha-specific genes HWP1, ALS1 and ALS3 were examined in the presence or absence of A. sativum, A. hirtifolium and A. cepaextracts by relative quantitative RT-PCR analysis.
Materials and Methods
Microorganisms
Ten clinical isolates of Candida albicans which were obtained from patients with systemic candidiasis and C. albicans ATCC 10231 as a reference quality-control strain was employed. All the isolates were maintained as sterile 20% (v/v) glycerol stocks and subcultured on Sabouraud dextrose agar with chloramphenicol (SDA, Difco Laboratories, Detroit, Michigan) at 35–37°C for 24–48 h to ensure viability and purity prior to testing.
Preparation of plant extracts
The bulbs of Allium sativum, Allium hirtifolium and Allium cepa were sourced from a Shiraz, Iran traditional medicine shop in mid-Mayand voucher specimens were confirmed byDepartment of Botany, University of Tehran. The plants were extracted according to the method described by Khodavandiet al. (2014a) with slight modifications. The fresh bulbs of A. sativum, A. hirtifolium and A. cepa were washed with sterile distilled water, sliced and dried in the oven for at least two days. The dried plant bulbs were crushed to fine powder, passed through an 80 mesh sieve and stored in a sealed plastic bag.Subsequently, one g sample of the plant bulbs powder was mixed with 10 mlof sterile distilled water, ethanol or methanol to give a stock solution of 100 mg/ml(w/v). The extracts were incubated at room temperature for 30 min and finally the extracts had undergone a sequential extractionusing soxhlet. The organic solvents were from the bulbs were filtered by Whatman No. 1 filter paper and the filtrate fractions were concentrated. Eventually, the different concentrations of tested extracts were made by dissolving in 5% DMSO and filter-sterilized (0.22-μm durapore, Millipore).
Antimicrobial susceptibility tests
The antifungal disk diffusion susceptibility test was performed by following method of Clinical and Laboratory Standards Institute (CLSI- M44-A) for yeast cell with slight modification. Briefly, C. albicansisolates were cultured on SDA and passaged at least twice to ensure viability and purity, and then incubated overnight at 37 ºC. Subsequently, five colonies that are more than 1mm in diameter of the overnight grown culture of C. albicansisolates were inoculated into 5 ml of sterile 0.85% saline and vortexed for 15 s. Cell densities were adjusted to1 ×106 to 5 × 106 yeast cells/ml using spectrophotometer at 530 nm wavelengths to achieve the turbidity equivalent to 0.5 McFarland standards. Hundred μl of the cell suspension was poured on SDA and kept at room temperature for 15 min to dry. The prepared tested extracts was added at a final concentration of 50, 60, 70, 80, 90 and 100 mg/ml to paper discs and placed on the agar surface. Serial tenfold dilution was carried out to make different concentrations of plant extracts. Discs containing fluconazole (Sigma-Aldrich, St. Louis, MO, USA) with the concentration of 5 mg/ml were maintained as positive controls. The plate was incubated at 37°C for 24 h and observed for inhibition zone.Three technical replicates were performed for each test and experiment was repeated at least two times (Khodavandiet al., 2011a; 2014a, b).
In order to determine the MIC of the plant extracts against C. albicans, the extracts were subjected to broth dilution antifungal susceptibility test.The broth microdilution antifungal susceptibility test was carried out using CLSI reference method for yeast (M27-A3). The test was carried out using 96-well U-bottom tissue culture microplates containing 100 µl/well of (SDB, Fluka, Germany). Hundred μL of the twofold dilution of the different concentrations of tested extracts were added to the wells. The antifungal agent, fluconazole was used as positive control. Yeast cells inocula were prepared (530 nm, abs 0.08–0.1) and diluted to reach a final concentration of 5 × 102 – 2.5 × 103 yeast cells/ml in the wells. Then the plates were incubated at 35°C for 24h. The endpoint was defined as the lowest concentration of each antifungal that caused 50% and 90% growth inhibition compared to control-growth (Khodavandiet al., 2011a; 2014a, b).
MFC values of tested extracts were determined for C. albicans isolates according to Canton et al.(2004). Briefly; MFCs were evaluated by transferring 0.1 ml from clear MIC wells following 24h of incubation onto Petri dishes containing SDA. The MFC was defined as the lowest extracts concentration that that killed ≥ 99.9% of yeast cells.
To evaluate the fungicidal effects of tested extracts against C. albicans time-kill assay was performed. Four ml of growing culture (106 cell/ml) of representative C. albicans isolates was added to the same volume of 1 × the MIC of extracts, and extract free medium was used as the growth control. Samples were obtained from each tube at predetermined time points of 0, 2, 6, 12, 24, 36 and 48h for viable colony counts. The viable counts were determined using the serial dilution method after incubation at 35°C for 24 h. Thelower limit of reproducibly quantifiable CFU according to these methods was 50 CFU/ml. All assays were conducted in in triplicates(Khodavandiet al., 2011b; 2014a, b).
Evaluation of quorum-sensing quenching activity
Quorum-sensing quenching activity of tested extracts on the C. albicans was performed by following method by Lim et al. (2009). Briefly, C. albicans ATCC 10231yeast cells were grown in Winge medium (0.4% glucose, 0.6% yeast extract) and placed into a shaking incubator at 25 °C overnight. The yeast cells were washed in phosphate buffer saline (pH 7.2) and adjusted to a cell density of 1 × 106 cells/ml in RPMI-1640 with L-glutamine (Sigma) supplemented with10% fetal bovine serum (Gibco, Invitrogen) and 100 U/ml penicillin-streptomycin (Gibco, Invitrogen).The cultures were incubated at 37 °C with 5% CO2 without and with aqueous plant tested extracts at different concentrations based on MIC (1/4× MIC, 1/2× MIC, 1× MIC and 2× MIC). The cultures were used for qualitative assay, as well as RNA extraction and quantitative RT-PCR analysis. We employed inverted light microscopy method as a qualitative assay to investigate the quorum-sensing quenching activity of tested extracts on the C. albicans at the time intervals of 0, 12 and 24 h.
RNA extraction and cDNA amplification
A suspension containing different concentrations of aqueous plant tested extracts and 1×106 cells/ml of C. albicans ATCC 10231were preparedas in the sample preparation for quorum-sensing quenching activity as explained above.The mixture was subsequently pelleted by centrifugation at 3000 rpm for 10 minand washed usingPBS at least three times.Total RNA was extracted using the RNeasy Mini Kit(Qiagen, Hilden, Germany) according to the manufacturer’s instructions with slight modifications. RNA quality and quantity were determined by formaldehyde-denaturing agarose gel electrophoresis and Nano Drop Spectrophotometer ND-1000 (NanoDrop Technologies Inc., Wilmington, DE). Any genomic DNA contamination was removed using DNase I (Promega, USA).Total RNA (0.5 µg) was copied into complementary DNA (cDNA) with Moloney-Murine Leukemia Virus (MMLV) reverse transcriptase and random hexamers ((Fermentas, USA) according to the manufacturer’s instructions. Each reaction was performed in three technical replications for each of three hyphal induction procedures with and without tested extracts.
Relative real time RT-PCR
albicanstranscription regulator gene, TUP1 and hypha-specific genes,HWP1,ALS1 and ALS3 genes were amplified from the synthesized cDNA with primers as described in Table 1.Moreover, actin was established as a house-keeping gene to normalize the dissimilar RNA concentrations during RNA extraction. Semi-quantitative RT-PCR was conducted using TMSYBR Green qPCR Master Mix (Fermentas, EU) via Bio-Rad MiniOpticonTM system (USA). The cycling conditionsincluded an initial step at 50◦C for 2 min; holding at 95◦C for 10 min, 40 cycles of denaturation at 95◦C for 15 s and subsequently annealing at 60◦C for 1 min. Eventually, the melting reaction was performed from 72–99◦C. Relative gene expression was quantified by the Pfaffl method (Khodavandiet al., 2011a).
Table 1: Oligonucleotide primers used for PCR.
| Gene | Primer sequence (5’→3′) | PCR product size (bp) | Reference |
| TUP1 | Forward: CTCTTGGCGACAGGTGCAG | 224 | (Toyoda et al., 2004) |
| Reverse: GTGGTGACGCCGTCTTCGA | |||
| HWP1 | Forward: TCAGTTCCACTCATGCAACCA | 99 | (Khodavandi et al., 2011a) |
| Reverse: AGCACCGAAAGTCAATCTCATGT | |||
| ALS1 | Forward: TTCTCATGAATCAGCATCCACAA | 53 | (Green et al., 2005) |
| Reverse:CAGAATTTTCACCCATACTTGGTTTC | |||
| ALS3 | Forward: AATGGTCCTTATGAATCACCATCTACTA | 56 | (Green et al., 2005) |
| Reverse:GAGTTTTCATCCATACTTGATTTCACAT | |||
| ACT | Forward: GAGTTGCTCCAGAAGAACATCCAG | 199 | (Lim et al., 2009) |
| Reverse:TGAGTAACACCATCACCAGAATCC |
Statistical analysis
All experiments were performed in triplicate. Results are expressed as mean value ± standard deviations (S.D) of three replicates and analyzed by using the software SPSS 20.0 for windows (SPSS Inc. Chicago, IL, USA).
Results
The results of preliminary disc diffusion assay screening on A. sativum, A. hirtifolium and A.cepaextractsrevealed that tested extractscould able to show the strong inhibitory activity against C. albicans isolates comparable to standard antifungal agent.The antimicrobial activity (assessed in terms of inhibition zone) of the tested extracts was found to be more potent at a concentration of 100 mg/ml(Table 2).However, A. hirtifoliumextract showed the strongest inhibitory zone against C. albicans isolates in comparison to A. sativum and A.cepa. The results obtained using three different solvent showed no significant differences of inhibition of tested extracts on the growth of C. albicans isolates.The MIC and MFC valuesof these tested extracts against C. albicans isolates are shown in Table 3.The MIC for all isolates was ≤ 25 mg/ml for tested extracts, with ranged from 5-15 mg/ml for A. hirtifolium, 11-25 mg/ml for A. sativum and A.cepa. These results indicated that the best antimicrobial activity was observed for A. hirtifoliumextract with lowest MIC value 5-8 mg/ml, MFC value 11-14 mg/ml and inhibition zone 27.55 ± 0.36. The killing patterns of extracts tested against C. albicans isolates were shown in Fig. 1. All tested extracts significantly reduced the number of viable cells at different time intervals (p≤ 0.0001).
Table 2.Antimicrobial activities of different Allium sativum, A. hirtifolium and A. cepa extracts against isolates of Candida albicansat a concentration of 100 mg/ml of tested extracts.
| Isolates | Inhibition zone (mm) | |||||||||
| A. sativum | A. hirtifolium | A. cepa | ||||||||
| Ethanol extract | Methanolextract | Aqueous extract | Ethanol extract | Methanolextract | Aqueous extract | Ethanol extract | Methanolextract | Aqueous extract | Fluconazole | |
| C. albicans ATCC 10231 | 17.15 ± 0.20 | 18.04 ± 0.11 | 17.12 ± 0.08 | 26.80 ± 0.01 | 25.00 ± 0.11 | 25.55 ± 0.90 | 17.03 ± 0.33 | 17.00 ± 0.10 | 16.03 ± 0.11 | 33.01 ± 0.01 |
| CI-1 | 17.20 ± 0.18 | 18.34 ± 0.16 | 16.90 ± 0.04 | 25.12 ± 0.34 | 25.65 ± 0.74 | 24.15 ± 0.44 | 17.56 ± 0.54 | 16.05 ± 0.11 | 14.22 ± 0.09 | 33.11 ± 0.16 |
| CI-2 | 17.15 ± 0.66 | 17.87 ± 0.68 | 16.70 ± 0.19 | 25.01 ± 0.21 | 24.85 ± 0.54 | 24.15 ± 0.44 | 16.32 ± 0.16 | 17.35 ± 0.24 | 15.15 ± 0.64 | 32.32 ± 0.51 |
| CI-3 | 16.37 ± 0.35 | 17.11 ± 0.46 | 16.96 ± 0.19 | 26.01 ± 0.18 | 25.01 ± 0.78 | 26.11 ± 0.10 | 16.17 ± 0.16 | 16.71 ± 0.31 | 14.57 ± 0.24 | 33.00 ± 0.09 |
| CI-4 | 17.29 ± 0.15 | 18.20 ± 0.33 | 15.80 ± 0.18 | 26.09 ± 0.30 | 25.19 ± 0.20 | 25.35 ± 0.30 | 17.05 ± 0.23 | 15.26 ± 0.44 | 13.28 ± 0.17 | 33.10 ± 0.11 |
| CI-5 | 16.33 ± 0.30 | 18.10 ± 0.06 | 17.10 ± 0.19 | 25.25 ± 0.22 | 24.45 ± 0.16 | 24.05 ± 0.12 | 15.15 ± 0.11 | 16.85 ± 0.02 | 14.75 ± 0.17 | 33.00 ± 0.03 |
| CI-6 | 16.62 ± 0.11 | 17.03 ±0.19 | 16.05 ± 0.24 | 26.18 ± 0.12 | 24.17 ± 0.14 | 26.14 ± 0.44 | 16.10 ± 0.11 | 17.11 ± 0.22 | 13.73 ± 0.45 | 32.02 ± 0.15 |
| CI-7 | 17.26 ± 0.08 | 17.94 ± 0.73 | 17.03 ± 0.18 | 25.22± 0.12 | 25.20 ± 0.10 | 24.00 ± 0.90 | 15.36± 0.16 | 15.20 ± 0.20 | 14.35 ± 0.30 | 33.11 ± 0.11 |
| CI-8 | 17.04 ± 0.25 | 18.41± 0.45 | 15.99 ± 0.18 | 25.65 ± 0.10 | 25.85 ± 0.80 | 25.35 ± 0.40 | 17.55 ± 0.18 | 16.05 ± 0.11 | 14.75 ± 0.68 | 33.01 ± 0.11 |
| CI-9 | 16.30 ± 0.05 | 17.65 ± 0.10 | 16.35 ± 0.12 | 25.27 ± 0.68 | 24.23 ± 0.48 | 26.33 ± 0.18 | 16.36 ± 0.10 | 15.23 ± 0.11 | 14.55 ± 0.48 | 33.07 ± 0.22 |
| CI-10 | 17.04 ± 0.40 | 17.76 ± 0.11 | 15.13 ± 0.25 | 26.45 ± 0.66 | 24.05 ± 0.16 | 27.55 ± 0.36 | 15.76 ± 0.26 | 15.11 ± 0.31 | 15.90 ± 0.66 | 32.22 ± 0.22 |
CI: Clinical isolates of C. albicans
Data are means ± standard deviation of three independent experiment
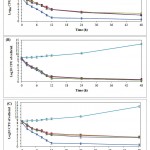 |
Figure 1: Time dependent killing of sessile CandidaalbicansATCC 10231 cells treated by different (A) Allium sativum, (B) A. hirtifolium and (C)A. cepa extracts.
|
Findings from the microscopic examination displayed quorum-sensing quenching activity of tested extracts on the C. albicansATCC 10231at the time intervals. C. albicans cells treated with A. hirtifolium,A. sativum and A.cepa extracts showed asignificant reduction in the number of yeast form. In addition, the transition from a yeast cell to the hyphal form was reduced, while the untreated control was able to form hyphae and structured to show the pre-biofilm after 24 h incubation (Fig. 2).
 |
Figure 2: Inverted light microscopic view of the quorum-sensing quenching activity of tested extracts on the Candida albicans ATCC 10231 at different time intervals. (A) Untreated control(B) Allium sativumethanol extract (C) A. sativummethanol extract (D) A. sativum aqueous extract(E) A. hirtifoliumethanol extract (F) A. hirtifoliummethanol extract (G) A. hirtifolium aqueous extract(H) A. cepaethanol extract (I) A. cepa methanol extract (J) A. cepaaqueous extract. Magnification × 40, Bar = 50 µm.
|
Relative quantitative RT-PCR analysis of transcriptional repressor and hypha-specific genes were conducted in C. albicansATCC 10231exposed to A. hirtifolium,A. sativum and A.cepa extracts. The expression levels of the transcriptional repressor and hypha-specific genes were significantly different (p≤ 0.0001) at all concentrations of tested extracts based on MIC(Fig. 3). In addition, patterns of C. albicans gene expression in response to quorum-sensing quenching activity of tested extracts were similar for hypha-specific genes HWP1,ALS1 and ALS3. While, the expression of TUP1 was different from the expression of HWP1,ALS1 and ALS3 as expected. The expression level of HWP1,ALS1 and ALS3 were significant (p≤ 0.0001) down-regulated compared to untreated control. In the present study, thetranscriptional repressor and hypha-specific genes demonstrated the highest fold change expression inC. albicans treated with A. hirtifolium compared to A. sativum and A.cepa extracts. The fold changes in terms of HWP1 expression to untreated control for 2× MIC, 1× MIC, ½× MIC and ¼× MIC of A. hirtifolium were 5.56 ± 0.002, 2.44 ± 0.001, 1.70 ± 0.002 and 1.22 ± 0.004, respectively. Moreover, the fold changes in untreated control in terms of ALS1 expression for 2× MIC, 1× MIC, ½× MIC and ¼× MIC of A. hirtifolium were 4.76 ± 0.005, 1.92 ± 0.002, 1.41± 0.004 and 1.14 ± 0.004, respectively. The ALS3 mRNA was down-regulated 4.17 ± 0.001, 2.13± 0.004, 1.70± 0.001 and 1.14 ± 0.003at A. hirtifolium concentrations of 2× MIC, 1× MIC, ½× MIC and ¼× MIC, respectively.While, the fold changes regarding to TUP1 expression for 2× MIC, 1× MIC, ½× MIC and ¼× MIC of A. hirtifolium were 2.81 ± 0.001, 2.59± 0.002, 2.35± 0.00 and 2.00 ± 0.002, respectively. The authenticity of the PCR products was verifiedby DNA sequencing method using an outsourcing sequencingservice. The sequences displayed high similarity analyzed via nucleotide BLAST in Gene Bank and confirmed in terms of homology to the related genes.
 |
Figure 3: Relative quantitation of (A) TUP1, (B) HWP1, (C) ALS1 and (D) ALS3 gene expression (normalized to house-keeping gene, actin) calculated with Log2 in Candida albicans ATCC 10231 after 24 h of treatment with different concentrations of tested extracts by real time RT-PCR. Data are means of fold changes with standard deviation from three independent experiments amplified in triplicates.
|
Discussion
Quorum-sensing quenching activity has been a promising strategy to combat microbial infections. In this way it does not impose any selection pressure on the pathogen and, as such, a predominantly resistant population should be less likely to emerge. Plant and their derived metabolites have been extensively studied as quorum-sensing quenching approaches(Adonizio et al., 2006; Koh and Tham, 2011; Koh et al., 2013; Viswanathan et al., 2015).These data demonstrate that some species of the genus Allium play an important role in quorum-sensing quenching activity. These findings are consistent with previous observations demonstrate that quorum-sensing quenching activitycould be induced by the genus Allium(Sharma and Jangid, 2013; Lade et al., 2014; Viswanathan et al., 2015).
The antimicrobial susceptibility results indicated that all C. albicans isolates were susceptible to plant extract tested. It is well known that the genus Alliumhave excellent inhibitory activity against C. albicans(Shuford et al., 2005; Palmeira-de-Oliveira et al.,2013; Vashisthet al., 2013; Khodavandi et al., 2014a).In this study, we analyzed the antifungal effects of A. sativum, A. hirtifolium and A. cepaextracts quantitatively and qualitatively by measuring the inhibition zone, minimum inhibitory concentration (MIC), minimal fungicidal concentration (MFC) and quorum-sensing quenching activity of C. albicans. In addition, we evaluated the expression analysis of selected genes related to quorum-sensing of C. albicans treated by A. sativum, A. hirtifolium and A. cepaextracts. The susceptibility profile exhibited for all C. albicans isolates to tested extracts. The genus Alliumhas been defined as the medications for many infectious diseases. It is demonstrated that the main bioactive components that originated from the genus Allium are sulfur-containing and non-sulfur components such as allicin and ajoene showing antifungal activities (Yoshida et al., 1987; Khodavandi et al., 2011a; Palmeira-de-Oliveira et al., 2013).
Quorum-sensing quenching activity of tested extracts on the C. albicanscompletely reduced the number of yeast form. This work demonstrated the ability of A. hirtifolium,A. sativum and A.cepa extracts to inhibit the transition from yeast to hyphae cells, a step essential to the virulence of Candida, suggesting that tested extracts could decrease the ability of C. albicans cells to cause disease. Similarly, Khodavandi et al. (2011c) found that the allicin damaged the cell surface of C. albicansresulting in the fungal load reduction and host survival timein alleviating systemic C. albicans infections.
In this study, the expression of HWP1,ALS1 and ALS3mRNA was found to be correlated with the transition from yeast to hyphae cells in C. albicans treated with A. hirtifolium,A. sativum and A.cepa extracts. The cells treated with tested extracts resulted in up-regulation of TUP1 mRNA expression, which could result in relief of transcriptional repression at hypha-related gene promoters. The down-regulatedhypha-specific genes HWP1,ALS1 and ALS3, consistent with the morphological yeast-to-hyphal switch which completely reduced the number of yeast form and inhibit the transition from yeast to hyphae cells. In addition to signal transduction pathways, C. albicans yeast and filamentous growth are controlled by an assortment of quorum-sensing molecules which trigger hyphal formation and biofilm development (Kruppa, 2008; Han et al., 2011; Sharma and Jangid, 2013). Previous investigations demonstrated that Tup1plays an important role in the repression of filamentous growth and the expression of hypha-specific genes in C. albicans (Braun et al., 2000; Kebaara et al., 2008). In addition, some important genes in C. albicans encode adhesin and invasion and contribute to biofilm formation. ALS family is included of several glycosylphosphatidylinositol (GPI)-linked cell surface glycoproteins. Of the Als proteins, the hypha-associated adhesin Als1 and Als3 is especially important for adhesion. Another important adhesin of C. albicans is Hwp1, which is a hypha-associated GPI-linked protein. Tup1 acts negatively to regulate filamentous growth. HWP1 and ALS formerly are activated by Efg1 and repressed by Tup1(Hoyer et al., 2008; Bastidaset al., 2009; Mayeret al., 2013).Quorum-sensing quenching activity has been reported from A. sativum, and it has been found that A. sativum extract has a preference for the genes belonging to virulence and pathogenesis of Pseudomonas aeruginosa including like LasA, LasB (coding for elastase and protease), rhlAB encoding rhamnolipid,chiC (encoding for chitinase), as well as aprA, phzA1B, phzS, phzC2D2E2F2G2, and PA1L (Kalia, 2013).
Table 3: Relative MIC and MFC values of different Allium sativum, A. hirtifolium and A. cepa extracts against isolates of Candida albicans.
| Tested extracts/Isolates | C. albicans ATCC
10231 |
CI-1 | CI-2 | CI-3 | CI-4 | CI-5 | CI-6 | CI-7 | CI-8 | CI-9 | CI-10 | ||
| A. sativuma | Ethanol
extract
|
MIC50 | 12-15 | 11-14 | 11-14 | 11-14 | 12-15 | 12-15 | 11-14 | 11-14 | 12-15 | 12-15 | 11-14 |
| MIC90 | 21-24 | 21-24 | 21-24 | 21-24 | 22-25 | 22-25 | 21-24 | 21-24 | 21-24 | 22-25 | 21-24 | ||
| MFC | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | ||
|
Methanol extract |
MIC50 | 11-14 | 11-14 | 11-14 | 11-14 | 11-14 | 12-15 | 12-15 | 11-14 | 12-15 | 11-14 | 11-14 | |
| MIC90 | 21-24 | 21-24 | 21-24 | 21-24 | 21-24 | 22-25 | 22-25 | 21-24 | 22-25 | 21-24 | 21-24 | ||
| MFC | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | ||
|
Aqueous extract |
MIC50 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | |
| MIC90 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | ||
| MFC | 24-27 | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | ||
| A. hirtifoliuma | Ethanol
extract
|
MIC50 | 6-9 | 5-8 | 6-9 | 6-9 | 5-8 | 5-8 | 5-8 | 5-8 | 5-8 | 5-8 | 5-8 |
| MIC90 | 11-14 | 10-13 | 11-14 | 11-14 | 10-13 | 10-13 | 10-13 | 10-13 | 10-13 | 10-13 | 10-13 | ||
| MFC | 13-16 | 11-14 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 11-14 | 12-15 | ||
| Methanol extract
|
MIC50 | 6-9 | 5-8 | 6-9 | 6-9 | 5-8 | 5-8 | 5-8 | 5-8 | 5-8 | 5-8 | 5-8 | |
| MIC90 | 11-14 | 10-13 | 11-14 | 12-15 | 10-13 | 10-13 | 10-13 | 10-13 | 10-13 | 10-13 | 11-14 | ||
| MFC | 13-16 | 13-16 | 12-15 | 13-16 | 13-16 | 13-16 | 11-14 | 12-15 | 12-15 | 12-15 | 12-15 | ||
| Aqueous extract | MIC50 | 5-8 | 5-8 | 6-9 | 6-9 | 6-9 | 6-9 | 5-8 | 6-9 | 5-8 | 6-9 | 6-9 | |
| MIC90 | 10-13 | 10-13 | 10-13 | 10-13 | 11-14 | 11-14 | 10-13 | 10-13 | 11-14 | 10-13 | 11-14 | ||
| MFC | 12-15 | 13-16 | 13-16 | 12-15 | 12-15 | 12-15 | 13-16 | 13-16 | 13-16 | 13-16 | 13-16 | ||
| A. cepaa | Ethanol
extract
|
MIC50 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 |
| MIC90 | 22-25 | 21-24 | 22-25 | 21-24 | 22-25 | 22-25 | 22-25 | 22-25 | 21-24 | 21-24 | 21-24 | ||
| MFC | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | ||
| Methanol extract
|
MIC50 | 12-15 | 11-14 | 12-15 | 11-14 | 11-14 | 12-15 | 12-15 | 11-14 | 11-14 | 11-14 | 12-15 | |
| MIC90 | 22-25 | 21-24 | 22-25 | 21-24 | 21-24 | 22-25 | 22-25 | 21-24 | 21-24 | 21-24 | 22-25 | ||
| MFC | 25-28 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | 24-27 | ||
| Aqueous extract | MIC50 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | 12-15 | |
| MIC90 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | 22-25 | ||
| MFC | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | 25-28 | ||
| Fluconazoleb | MIC50 | 0.5-2 | 0.5-2 | 4-8 | 1-4 | 1-4 | 1-4 | 1-4 | 4-8 | 4-8 | 1-4 | 1-4 | |
| MIC90 | 1-4 | 1-4 | 16-32 | 8-16 | 8-16 | 8-16 | 8-16 | 16-32 | 16-32 | 8-16 | 8-16 | ||
| MFC | 8-16 | 8-16 | 32->64 | 16-32 | 16-32 | 16-32 | 16-32 | 32->64 | 32->64 | 16-32 | 16-32 | ||
amg/mL
bμg/mL
CI: Clinical isolates of C. albicans
Data are means ± standard deviation of three independent experiments
Conclusion
Whether these events reflect the potential of A. hirtifolium,A. sativum and A.cepa extracts for quorum-sensing quenching activity in C. albicans which differentially expresses specific genes, requires further dissection. In summary, these results link the transcriptional repression gene to regulation of hypha-specific genes in a major human fungal pathogen. These functional clues will be combined with information from other approaches to further define of quorum-sensing quenching mechanisms.This might provide useful information to develop powerful new therapeutic approaches.
Acknowledgements
Thanks are due to Islamic Azad University of Gachsaran.
References
- Adonizio, A.L., Downum, K., Bennett, B.C., Mathee, K. Anti-quorum sensing activity of medicinal plants in southern Florida. J Ethnopharmacol 2006;105: 427-435.
CrossRef - Bastidas, R.J., Heitman, J., Cardenas, M.E. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLOS Pathog 2009;5,
- Braun, B.R., Head, W.S., Wang, M.X., Johnson, A.D. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics2000; 156: 31-44.
- Canton, E., Peman, J., Gobernado, M., Viudes, A., Espinel-Ingroff, A. Patterns of amphotericin B killing kinetics against seven CandidaAntimicrob Agents Chemother 2004; 48: 2477-2482.
- Cuellar-Cruz, M., Lopez-Romero, E., Villagomez-Castr, J.C., Ruiz-Baca, E. Candida species: new insights into biofilm formation. Future Microbiol2012; 7: 755-771.
CrossRef - da Silva-Rocha, W.P., Lemos, V.L.B., Svidizisnki, T.I.E., Milan, E.P., Chaves, G.M.Candida species distribution, genotyping and virulence factors of Candida albicans isolated from the oral cavity of kidney transplant recipients of two geographic regions of Brazil. BMC Oral Health2014; 14: 20.
CrossRef - Green, C.B., Zhao, X., Yeater, K.M.,Hoyer, L.L. Construction and real-time RT-PCR validation of Candida albicans PALS-GFP reporter strains and their use in flow cytometry analysis of ALS gene expression in budding and filamenting cells. Microbiology 2005; 151: 1051-1060.
CrossRef - Han, T.L., Cannon, R.D., Villas-Bôas, S.G. The metabolic basis of Candida albicans morphogenesis and quorum sensing. Fungal Genet Biol 2011;48: 747-763.
CrossRef - Hoyer, L.L., Green, C.B., Oh, S.H., Zhao, X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family-a sticky pursuit. Med Mycol 2008; 46: 1-15.
CrossRef - Kalia, V.C. Quorum sensing inhibitors: an overview. Biotechnol Adv 2013; 31: 224-245.
CrossRef - Kebaara, B.W., Langford, M.L., Navarathna, D.H.M.L.P., Dumitru, R., Nickerson, K.W., Atkin, A.L. Candida albicans Tup1 is involved in farnesol-mediated inhibition of filamentous-growth induction.Eukaryotic cell2008;7: 980-987.
CrossRef - Khodavandi, A., Alizadeh, F., Aala, F., Sekawi, Z.,Chong, P.P. In vitro investigation of antifungal activity of allicin alone and in combination with azoles against Candida Mycopathologia2010; 169: 287-295.
- Khodavandi, A., Harmal, N.S., Alizadeh, F., Scully, O.J., Sidik, S.H.M., Othman, F., Sekawi, Z., Ng, K.P., Chong, P.P. Comparison between allicin and fluconazole in Candida albicans biofilm inhibition and in suppression of HWP1 gene expression. Phytomedicine2011a; 19: 56-63.
- Khodavandi, A., Alizadeh, F., Harmal, N.S., Sidik, S.M., Othman, F., Sekawi, Z.,Chong, P.P. Expression analysis of SIR2 and SAPs1-4 gene expression in Candida albicans treated with allicin compared to fluconazole. Tropical Biomedic 2011b; 28: 589-598.
- Khodavandi, A., Alizadeh, F., Harmal, N.S., Sidik, S.H.M., Othman, F., Sekawi, Z., Farbood-niaye Jahromi, M.A., Ng, K.P.,Chong, P.P. Comparison between efficacy of allicin and fuconazole against Candida albicansinvitro and in a systemic candidiasis mouse model.FEMS Microbiol Lett 2011c; 315: 87–93.
CrossRef - Khodavandi, A., Alizadeh, F., Namvar, F., Rosfarizan, M., Chong, P.P. Anti-Candida potential of Allium ascalonicum Linn: antibiofilm activity and biomolecular mechanism of action. J Pure Appl Microbiol 2014a; 8: 349-356.
- Khodavandi, A., Alizadeh, F., Aghai Vanda, N., Karimi, G.,Chong, P.P. Possible mechanisms of the antifungal activity of fluconazole in combination with terbinafine against Candida albicans. Pharm Biol 2014b; 52: 1505-1509.
CrossRef - Koh, K.H., Tham, F.Y. Screening of traditional Chinese medicinal plants for quorum-sensing inhibitors activity.J Microbiol Immunol Infect 2011; 44: 144-148.
CrossRef - Koh, C.L., Sam, C.K., Yin, W.F., Tan, L.Y., Krishnan, T., Chong, Y.M., Chan, K.G. Plant-derived natural products as sources of anti-quorum sensing compounds. Sensors2013; 13: 6217-6228.
CrossRef - Kruppa, M. Quorum sensing and Candida albicans. Mycoses2008; 52: 1-10.
CrossRef - Lade, H., Paul, D., Kweon, J.H. Quorum quenching mediated approaches for control of membrane biofouling. Inter J Biol Sci 2014; 10: 550-565.
CrossRef - Lim, C.S.Y., Wong, W.F., Rozita, R., Ng, K.P., Seow, H.F., Chong, P.P. 2-dodecanol (decyl methyl carbinol) inhibits hyphal formation and SIR2 expression in albicans. J Basic Microbiol 2009; 49: 579-583.
CrossRef - Mayer, F.L., Wilson, D., Hube, B. Candida albicans pathogenicity mechanisms.Virulence2013; 4: 119-128.
CrossRef - Nett, J.E. Future directions for anti-biofilm therapeutics targeting Candida. Expert Rev Anti Infect Ther 2014; 12: 375-382.
CrossRef - Odds, F.C., Brown, A.J.P., Grow, N.A.R. Antifungal agents: mechanisms of action.TRENDS Microbiol 2003; 11: 272-279.
CrossRef - Palmeira-de-Oliveira, A., Silva, B.M., Palmeira-de-Oliveira, R., Martinez-de-Oliveira, J., Salgueiro, L. Are plant extracts a potential therapeutic approach for genital infections?Curr Med Chem 2013; 20: 2914-2928.
CrossRef - Sharma, R., Jangid, K. Fungal quorum sensing inhibitors. In: Quorum Sensing vs Quorum Quenching: A Battle with no End in Sight. Kalia, V.C. (editor), 1th edition, New York, Springer, 2015, pp. 237-257.
CrossRef - Shuford, J.A., Steckelberg, J.M., Patel, R. Effects of fresh garlic extract on Candida albicans Antimicrob Agents Chemother 2005; 49: 473.
CrossRef - Toyoda, M., Cho, T., Kaminishi, H., Sudoh, M., Chibana, H. Transcriptional profiling of the early stages of germination in Candida albicans by real-time RT-PCR. FEMS Yeast Res 2004; 5: 287-296.
CrossRef - Vashisth, P., Nikhil, K., Pemmaraju, S.C., Pruth, P.A., Mallick, V., Singh, H., Patel, A., Mishra, N.C., Singh, R.P., Pruthi, V. Antibiofilm activity of quercetin-encapsulated cytocompatible nanofibers against Candida albicans. J Bioactive Compatible Polymers 2013; 28: 652-665.
CrossRef - Viswanathan, P., Rathinam, P., Suneeva, S.C. Plant quorum sensing inhibitors: food, medicinal plants, and others. In: Quorum Sensing vs Quorum Quenching: A Battle with no End in Sight. Kalia, V.C. (editor), 1th edition, New York, Springer, 2015,pp. 269-281.
CrossRef - Yoshida, S., Kasuga, S., Hayashi, N., Ushiroguchi, T., Matsuura, H., Shizutoshi, N. Antifungal activity of ajoene derived from garlic. Appl Environ Microbiol 1987; 53: 615-627.

This work is licensed under a Creative Commons Attribution 4.0 International License.





