How to Cite | Publication History | PlumX Article Matrix
Mahshid Mousavi1, Abbas Akhavan Sepahi1 and Taher Nejadsattari2
1Department of Microbiology, North Tehran branch, Islamic Azad University, Tehran, Iran.
2Department of Biology, Tehran Science and Research Branch, Islamic Azad University, Tehran, Iran.
Corresponding Author E-mail: akhavansepahy@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2339
ABSTRACT: Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous contaminants in the environment with potential mutagenicity and carcinogenicity. PAH bioremediation is considered as an effective and environmentally benign cleanup technology. In this study we isolated Bacillus sp.in soil samples with potential of naphthalene degradation and compared Bacillus spp. consortium and pure cultures biodegradation rate of naphthalene.Ten soil samples were collected from Tehran and around Tehran and Bacillus strains were isolated and identified by biochemical and molecular methods. After that, the amount of biosurfactant production was measured and emulsification test was conducted. Eventually, GC analysis was conducted on the consortium for naphthalene biodegradation investigation. Optimized growth conditions were also determined involved in degradation (nah genes). At the end, catabolic genes were identified. The isolates contained Bacillus cereus, Paenibacillus lactis, Bacillus fusiformis and Bacillus subtilis. All of the isolates had surface tension amount less than 40(mN/m) and growth in blood agar medium. It shows that all of them can produce biosurfactant, effectively.The surface tension amount for consortium was lower than pure cultures. In addition, the consortium showed the ability to tolerate 1000 ppm naphthalene concentration in medium and the maximum growth amount was in pH=6, 200 ppm naphthalene concentration, 150 rpm of shaker speed and yeast extract as an additional nitrogen source.The obtained results revealed that consortium of four isolates had more ability in naphthalene degradation as shown in other studies.
KEYWORDS: Naphthalene; Bacillus sp.; Consortium, Bioremediation
Download this article as:| Copy the following to cite this article: Mousavi M, Sepahi A. A, Nejadsattari T. Investigation on Naphthalene Biodegradation Ability in a Bacillus sp. Consortium Isolated from Soil Samples. Biotech Res Asia 2016;13(3). |
| Copy the following to cite this URL: Mousavi M, Sepahi A. A, Nejadsattari T. Investigation on Naphthalene Biodegradation Ability in a Bacillus sp. Consortium Isolated from Soil Samples. Biotech Res Asia 2016;13(3). Available from: https://www.biotech-asia.org/?p=15878 |
Introduction
Now a days, crude oil remains a major source of energy and chemical raw material. Large-scale production, transport, use and disposal of petroleum globally have made it a major contaminant in both prevalence and quantity in the environment1.
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous oil contaminants in the environment with potential mutagenicity and carcinogenicity. They are generated from natural combustion processes as well as from human activities2. Industrialization & accidental spillage have increased the pollution of Hydrocarbon compounds in the soil as well as water 3. Among the released compounds followings are of major concern: biphenyl, polychlorinated biphenyls (PCBs), petroleum chemicals including (PAHs) and BTEX (benzene, toluene, ethylbenzene and xylenes), pesticides, dioxins, furans, and flame retardants4.
Naphthalene, a hydrocarbon with two fused aromatic rings, the first member of PAH group and one of the 16 PAHs is classified as priority pollutants by Environmental Protection Agency (EPA) of United States, is a frequent pollutant established in nature 5. Naphthalene is an abundant component of coal tar. Also petroleum commonly contains naphthalene. Naphthalene is a raw chemical for industrial syntheses, in particular of phthalic anhydride. Human exposure to naphthalene was formerly not uncommon due to its (now obsolete) use as a moth repellent. Naphthalene at high concentration may cause hemolytic anemia and some other conditions, whereas the tumorigenic potential is presently considered low 6. Naphthalene has long been used as a model compound in PAH bioremediation studies.
PAH bioremediation is considered as an effective and environmentally benign cleanup technology as it involves the partial or complete bioconversion of these pollutants to microbial biomass, carbon dioxide and water7. Also, bioremediation has low cost and limited impact on the environment2. The extent and rate of biodegradation depends on many factors including bioavailability of the carbon substrate, pH, temperature, oxygen, microbial load and strain, degree of acclimation, accessibility of nutrients, chemical structure of the compound, cellular transport properties, and chemical partitioning in growth medium [5,8]. A variety of bacteria have shown the potential of organic pollutants biodegradation, such as Arthrobacter spp., Bacillus spp., Pesaudomonas spp. and Mycobacterium spp. 9.
Microbial consortia have been widely used in cleanup of a number of pollutants in laboratory and field bioremediation studies. It is generally thought that microbial consortia are more effective than pure cultures in biodegradation of PAHs. This is possibly because of the broader enzymatic capacity achieved and the formation of toxic intermediate metabolites is counteracted by the selection of these dead-end products formed mainly by co-metabolism processes 5.
The aim of this study was to isolate Bacillus sp. from soil samples, test their ability as aconsortium to degrade naphthalene in liquid medium and compare it to pure cultures.
Materials and Methods
Sample Collection
Soil samples were collected from ten sites in Tehran andaround Tehran. The samples were collected in pre-sterilized sample bottles and the samples duly labeled and stored at 4 o C for further analysis.
Chemicals
Naphthalene, culture medium powders and mineral salts were purchased from Merck® chemical company.
Isolation and characterization of crude oil– degrading Bacillus sp
The soil samples were passed through a sieve to remove large debris pieces and vegetation.1 g of each soil sample was dissolved in 9 mL of distilled water and heated at 80°C for 20 minutes. The samples were cultivated in 50 ml MSM medium with naphthalene concentration of 50 ppm for two weeks. At the end of each week, the colonies were isolated by pour plate technique on nutrient agar and MSM agar and incubated at 37 oC for 48 h. [10].The developed colonies were counted in plates and the average number of colonies was determinedper plates. The number of total bacteria (CFU) was determinedper gram dry weight of soil. The composition of the MSM medium used in this study was as follows (g/L):
NaNO3 (2.0 g/L), NaCl(0.8 g/L), KCl (0.8 g/ L),CaCl2.2H2O(0.1 g/L), KH2PO4(2.0 g/L), Na2HPO4.12 H2O(2.0 g/L), MgSO4(0.2 g/L), FeSO4.7H2O (0.001 g/L); 2 mL trace element stock solution composed of (g/L): FeCl3.6H2O(0.08 g/L), ZnSO4.H2O(0.75 g/L),COCl2.6H2O(0.08g/L),CuSO4.5H2O(0.075 g/L), MgSO4. H2O (0.75 g/L), H3BO3 (0.15 g/L), Na2MoO4. 2H2O (0.05 g/L). The initial pH was adjusted at 6.811.
Individual cultures were preliminarily identified by morphological and biochemical techniques using the taxonomic scheme of Bergey’s Manual of Determinative Bacteriology. Then, isolated bacteria were identified by molecular method as well.
16SrRNA sequencing method was used for identification. Universal primers were used for PCR (Polymerase Chain Reaction):
27F: 5′-AGTTTGATCCTGGCTCAG-3′
1495R: 5′-CTACGGCTACCTTGTT-3′
The reaction mixture (25 µL) contained: each primer at a concentration of 0.5 µL, 12.5 µL of master mix, 1 µL of DNA and 10.5 µL of double distilled water. The following thermal profile was used for the PCR (My Cycler, Bio-Rad Laboratories, and United States): 94 oC, 2min; 35 cycles of 94oC, 1 min; 55 oC, 1min; 72 oC, 2 min; 1 cycle of 72 oC, 10 min12. The samples were sent to Zist Takapou Company for sequencing. Then we used BLAST (Basic Local Alignment Search Tool) algorithm to compare the sequences.
Surface Tension Measurement
The isolated Bacillus cultivated on Blood Agar and incubated at 30°C for 72h. The bacteria with β haemolytic activity were selected for measuring the surface tension. The surface tension was measured using the Du Neouy ring method by tensiometer (KRUSS GmbH, Germany). All the measurements have done three times 13.
Emulsification test
This test was conducted and measured by using Cooper and Goldenberg method: 4 mL of Mineral Salt Medium after 48h incubation, and 6 mL hydrocarbon (oil) was added to each test tube. The mixture was vortexed at high speed for 2min, and then the test tubes were leaved to stand for 24h. The E24 index is given as percentage of height of emulsified layer (cm) divided by total height of the liquid column (cm) Cooper and Goldenberg 11.
Determination of Growth Curve
The strains were cultivated in nutrient brothmedium and placed on shaker (K5501, IKA®-Works,Germany) with 150 rpm shaking speed. O.D600nm of the medium was taken by using spectrophotometer (T90 UV-Vis,PG instruments Ltd, England) after every 12 hour for 8 days and the growth curve was depicted.
The Ability of Growth in Different Naphthalene Concentrations
The naphthalene concentration in culture medium was gradually increased up to 1000 ppm. O.D600nm of the medium was measured every day for a period of nine days and the naphthalene tolerance chart was drawn.
The Optimization of Growth Conditions
To maximize naphthalene degradation, parameter optimization was done using the Taguchi experimental design. The experimented parameters were PH (2, 4, 6, 8), shaking speed (100,120,150,200), nitrogen source (ammonium sulfate, sodium nitrate, yeast extract, peptone) and naphthalene concentration of the medium (50, 100, 150, 200 ppm). After this experiment surface tension of consortium was measured again.
G.C. Analysis
For GC (Gas Chromatography) analysis, the culture medium was first transferred to a 250 ml pre-ash conical flask by addition of 20 μL m-terphenyl as internal standard (prepared in acetone with concentration of 1000 mg.L−1) and 25 ml ethyl acetate and shacked for 15 min. After separation from the water layer, the organic solvent layer was transferred to a clean conical flask before the second extraction by another aliquot of 25 ml ethyl acetate. Finally, two extracts were combined together, dried by anhydrous Na2SO4 and the volume was adjusted to 50 ml. About 1 ml of the extract sample was transferred into a 1.5 ml brown vial and analyzed by GC-FID. Three vials were prepared, including control, fourth day and eighth day samples. .The GC-FID was equipped with a HP-5MS fused silica capillary column (60 m × 0.25 mm ID × 0.25 lm thicknesses, Agilent Technologies, USA) with the injector and detector temperature of 280°C and 300°C, respectively. Helium was used as the carrier gas. The oven temperature program was set as follows: The oven temperature program was from 80°C (for 2 min) to 120°C at a rate of 10°C/min and from 120°C to 300°C at a rate of 4°C/minute and held at 300°C for 15 min. The identification and quantification of chemicals were conducted based on matching their retention times of standards14.
Identification of catabolic genes
To reveal the proportion of colonies hybridizing for each probe, dot blots were prepared with 3 µg of genomic DNA from each isolate applied to a positively charged nylon membrane. Southern blot consisting of 10 Wl of PCR products amplified from DNA extracted directly from each soil sample and resolved on an agarose gel, were also blotted to a positively charged nylon membrane for analysis by hybridization with the corresponding 32P-labelled probes.
PCR amplification was carried out in 20 mM TrisHCl (pH 8.4); 50 mM KCl; 1.25 mM MgCl2; dNTPs at 200 WM each, 2.5 units PLATINUM Taq DNA polymerase; 0.2 µM forward and reverse primers; and template DNA at 0.1 µg per 50 µl reaction. The cycling conditions (Techne Cyclogene Thermal Cycler) were 5 min at 94³C, followed by 25 cycles (40 cycles when amplifying from DNA isolated from soil) of 94 oC for 2 min, 55 oC (nah primers) for 1 min, 72³C for 1 min, maximal ramp rates throughout, with the final 72 oC segment of the cycle extended to 10 min beforecooling to 4 oC. PCR products amplified from genomic DNA of PAH-degrading isolates were visualized by agarose gel electrophoresis15.
Statistical Analysis
We used one way ANOVA test for statistical analysis, also Taguchi experimental design was analyzed by factorial ANOVA,at a level of significance of p< 0.05.
Results
Isolation and identification
Four morphologically different Bacillus colonies were isolated from the samples. The maximum numbers of bacteria 4.1* 104 and the minimum numbers of PAHs degrading bacteria 1.8*104 of view.
The isolates were gram positive, rode shaped and had endospores. Figure 1 shows gram and malachite green stains’ results. The results of biochemical tests have shown in table1.
 |
Figure 1: a- Gram stain, b-Malachite green stain of isolates
|
The strains a, b, c and d were identified as Bacillus cereus, Paenibacillus lactis, Bacillus fusiformis and Bacillus subtilis respectively by 16SrRNA sequencing method. A 100bp marker was used. Figure 2 shows the band positions that indicated in the gel
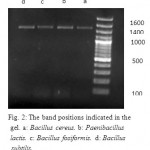 |
Figure 2: The band positions indicated in the gel. a: Bacillus cereus. b: Paenibacillus lactis. c: Bacillus fosiformis. d: Bacillus subtilis
|
Surface Tension Experiments
All fourisolates have shown colonies on Blood agarmedium, therefore they were selected for surface tension experiment.
The initial surface tension was 70 (mN/m), which was reduced to 38,33,35,37 and 29 (mN/m) by Bacillus cereus, Paenibacillus lactis, Bacillus fosiformis and Bacillus subtilis and consortium of four isolates respectively.
Emulsification test
The ability of emulsifying for the consortium was 85% and for Bacillus cereus, Paenibacillus lactis, Bacillus fosiformis and Bacillus subtilis were 76%,73%,77% and 79% respectively.
Growth Curve Study
Figure 3 shows the growth curve of the isolates in nutrient broth medium. Generation time for the isolates was 24 hours.
Naphthalene Toleration by Consortium
According to results, the consortium had ability to biodegrade 1000 ppm of naphthalene concentration in MSM liquid medium (Figure4). Growth on MSM agar medium after nine days confirmed this result.
Table1: Biochemical Tests’ Resultsfor Bacillus sp Isolated from Soil of Tehran and around Tehran
| Bacillus cereus | Paenibacillus lactis | Bacillus fosiformis | Bacillus subtilis | ||||||
| MR | + | – | + | + | |||||
| VP | – | + | – | – | |||||
| Simmon citrate | + | + | + | + | |||||
| Gelatinase | + | + | – | + | |||||
| Catalase 3% | + | + | + | + | |||||
| Catalase 10% | + | + | + | + | |||||
| Glucose | + | + | + | + | + | + | + | + | |
| anaerobic | aerobic | ||||||||
| Mannitol | + | + | – | – | + | + | + | + | |
| anaerobic | aerobic | ||||||||
| Xylose | + | + | – | – | + | + | + | + | |
| anaerobic | aerobic | ||||||||
| Arabinose | + | + | – | – | + | + | + | + | |
| anaerobic | aerobic | ||||||||
| SIM | ++- | – + – | ++- | ++- | |||||
| TSI | Alk/A,G | A/A | Alk/A,G | Alk/A,G | |||||
| Amylase | + | + | – | + | |||||
| Lecithinase | – | – | + | + | |||||
| Caseinase | + | – | – | – | |||||
Optimization Experiment
The results of optimization experiment showed that the optimumpH for consortiumgrowth was in MSM medium; also the best shaking speed for the growth was 150rpm. The best additional nitrogen source was yeastextract and naphthaleneconcentration at 200ppm had the best effect on growth. (Figure 5) And surface tension amountwas 26 (mN/m)in this culture.
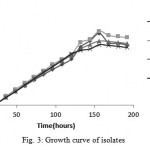 |
Figure 3: Growth curve of isolates
|
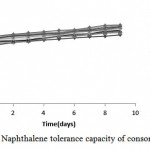 |
Figure 4: Naphthalene tolerance capacity of consortium
|
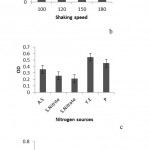 |
Figure 5: a. The comparison of consortium growth rate in different pH, b, and the comparison of consortium growth rate indifferent shaking speeds, c; the comparison of consortium growth rate in different extranitrogen sources. A.S: Ammonium sulfate. S. Nitrite: Sodium nitrite. S. Nitrate: Sodium nitrate. Y.E: Yeast extract and P: Peptone, d. the comparison of consortium growth rate in different concentrations of naphthalene
|
G.C. Analysis
In order to determine naphthalene degradation and intermediate metabolites formation we used GC analysis. The chromatograms were shown in Figure 6.
Identifications of Catabolic genes
All of naphthalene degrading isolates possessed nah genes.
Statistical Analysis
There was a significant difference 0.05 between the average of the bacteria in medium with and without PAHs. Also data was significantfor surface tension and optimization experiment.
Discussion
Isolation of Bacillus sp
In this study, ten soil samples were collected from Tehran and around Tehran. Five samples were from contaminated sites and five samples were from uncontaminated sites. Three of the contaminated samples contained Bacillus naphthalene degraders and one of the samples from uncontaminated sites contained Bacillus degraders, because the ability of native microbial population in biodegradation of contaminants were shown in former studies11.
According to the previous studies Bacillus cereus and Bacillus subtilis were isolated from soil samples which could degrade naphthalene and other PAHs, but to the best of our knowledge, Paenibacillus lactis with degradation abilities hasn’t isolated from soilsbefore11,16,17.
Surface Tension Measurement and Emulsification Test
The surface tension amount for consortium was the lowest; therefore it had the ability to produce more amount of biosurafctant than pure cultures and biodegrade naphthalene more effectively.
For the first time Banat in 1993 performed both haemolytic and surface tension experiment to determine biosurfactant production. Also in other studies hemolytic and surface tension experiment were used on Bacillus subtilis10,18. We selected the isolates with surface tension amount lower than 40 (mN/m), which contained all of strains. The consortium showed the lowest surface tension. That means it produced the maximum amount of biosurfactant 19. Akhavan etal in 2008 used emulsification test for Bacillus spp.11
 |
Figure 6: a. Chromatogram of solvent (ethyl acetate), b. Chromatogram of control sample, c. Chromatogram of sample collected from fourth day after growth, d. Chromatogram of sample collected from eight days after growth.
|
Growth Curve
The isolates had almost steady growth for the first day (Lag phase). After that, the growth rate was exponentially increased (Log phase)for the next six days. At the end, the growth rate was remained steady (Stationary phase). Growth curves of four isolates were approximately similar.
In 2012 a research conducted by kafilzadeh and growth curve of Pseudomonas sp. and Corynebacterium sp. on pyrene, phenantherene and naphthalene was studied and it involves all stages we saw in this study 20.
Growth in High Concentrations of Naphthalene
Figure 4 suggests an effective degradation of consortium and ability to adapt with naphthalene concentrations up to 1000ppm.
The Optimization study for consortium
The results of a previous study showed that the best nitrogen source for Bacillus growth is yeast extract 21. Also pH=7 was shown to be the best amount of pH for Bacillus fusiformis growth in the medium22.
G.C. Analysis
Figure 6 showed that naphthalene concentration in MSM liquid medium was reduced in eight days and intermediate metabolites from its degradation (like 1,2-dihydroxy 1,2-dihydro naphthalene) were increased.
This method was used to determine alkanes’ degradation by bacteria in India. The results showed that Bacillus subtilis is the best degrader23. In another study, itwas used on phenantherene and fluorene degradation by a consortium enriched from mangrove sediments14.
Identifications of Catabolic genes
Lloyd-Jones etal investigated catabolic genes involved in naphthalene and phenantherenein soils of New Zealand. nah genes were identified in Pseudomonas spp.15.
The Comparison of Pure Cultures and Consortium in Naphthalene Biodegradation
According to the results of this study the consortium of four isolates could biodegrade naphthalene more effectively than each culture. In a former study, a consortium of Bacillus spp. reduced surface tension of medium from 72 (mN/m) to 28 (mN/m), therefore it was more effective than pure cultures24. In another study, the consortium showed more ability for biodegradation of 2,4-dichlorophenol 16.Results of another study showed that a consortium of Psedumonas, Bacillus and Staphylococcus has more capability for biodegradation than pure cultures 25. Abdel et al in 2008 got the same results in their study on a Bacillus consortium.26
Conclusion
In this study we isloated Bacillus sp from soil samples and measured naphthalene biodegradation ability on pure cultures and their consortium. Results of phenotypic and genotipic identification showed that these isolates wereBacillus cereus, Paenibacillus lactis, Bacillus fusiformis and Bacillus subtilis with 99% of similarity index.The results of surface tension experiment showed that the consortium of four isolates produce more amount of biosurfactant and in emulsification test we understand the consortium was a better emulsifier. Therefore it was more effective in naphthalene biodegradation in comparison to pure cultures. The findings of this study suggest a potential bioremediation role of this Bacillus consortium in the removal of naphthalene from contaminated soils.
References
- Wang, H., Xu, R., Li, F., Qiao, J.,Zhang, B.Efficient degradation of lube oil by a mixed bacterial consortium. JESC., 2010;22:381-388.DOI: 10.1016/S1001-0742(09)60119-4
CrossRef - Mao, J., Luo, Y., Teng, Y.,Li, Z.Bioremediation of polycyclic aromatic hydrocarbon-contaminated soil by a bacterial consortium and associated microbial community changes. J Int Biod.,2012; 70:141-147.
DOI:10.1016/j.ibiod.2012.03.002.
CrossRef - Gomare, K.S., Lahane, M.N. Degradation of Polyaromatic Hydrocarbons by Isolated Cultures From Contaminated Soils at Petrol Pump Stations.IJRTSAT., 2011;1:9-13.
- Uhlik, O., Wald, J., Strejcek, M., Musilova, L., Ridl, J. et al..Identification of Bacteria Utilizing Biphenyl, Benzoate, and Naphthalene in Long-Term Contaminated Soil. PLoSone., 2012; 7:1-10.DOI:10.1371/journal.pone.0040653.
CrossRef - Raïssa, K.R., Augustin, M., Martin-Benoît, N.Naphthalene biodegradation by microbial consortia isolated from soils in Ngaoundere (Cameroon).IJES.,2012;3(1):1-10.DOI:10.6088/I jes.2012030131001.
- Musat, F., Galushko, A., Jacob, J., Widdel, F.,Kube, M.Anaerobic degradation of naphthalene and 2-methylnaphthalene by strains of marine sulfate-reducing bacteria. EnvMic.,2009;11:209-219.
CrossRef - Pawar, A.N., Ugale, S.S., More, M.G., Kokani, N.F., Khandelwal, S.R. Biological Degradation of Naphthalene: A New Era. J Bioremed Biodeg., 2013; 4:1-5. DOI: 104172/2155-6199.1000203.
- Dou, J., Shuairan, L., Cheng, L., Ding, A., Liu, X., Yun, Y.The Enhancement of Naphthalene Degradation in Soil by Hydroxypropyl-ȕ-cyclodextrin.Pro Env Sci., 2011;10:26-31. DOI:10.1016/j.proenv.2011.09.006.
CrossRef - Seo, J-S., Keum, Y-S.,Li, Q.X.Bacterial Degradarion of Aromatic Compounds.IJERPH.,2009;6:278- 309. DOI:10.3390/ijerph6010278.
CrossRef - Banat I. The isolation of thermophilic biosurfactant producing bacteria.Biotechnollett.,1993;15: 591-594.
CrossRef - Akhavan Sepahi, A., Dejban Golpasha, I.,Emami, M.,Nakhoda, A.M.Isolation and characterization of crude oil degradingBacillus spp.Iran. J. Environ. Health. Sci. Eng. 2008;,5:149-154.
- Assareh, R., Shahbani Zahiri, H., Akbari Noghabi, K., Aminzadeh, S.,Bakhshi Khaniki, G.Characterization of the newly isolated Geobacillus sp. T1, the efficient cellulase producer on untreated barley and wheat straws. Bioresource Technol.,2012; 120:99-105.DOI:10.1016/j.biortech.2012.06.027.
CrossRef - Banat I. M. Biosurfactant production and possible uses in Microbial Enhanced Oil Recovery and Oil Pollution Remediation.Bioresource Technol.,1995;51: 1-12.
CrossRef - Shahriari Moghadam, M., Ebrahimipour, G.H., Abtahi, B., Ghassempour, A.,Seyed Hashtroudi, M. Biodegradation of polycyclic aromatic hydrocarbons by a bacterial consortium enriched from mangrove sediments.IJEHSE.,2013;12:1-9.
- Lloyd-Jons, G., D.Laurie, A., W.F. Hunter, D.,Fraser, R. Analysis of catabolic genes for naphthalene and phenanthrene degradation in contaminated New Zealand soils. FEMSMicEco.,1999;29: 69-79.
CrossRef - Herrera, Y., Okoh, A. I.,Alvarez, L. R.Biodegradation of 2,4-dichlorophenol by a Bacillus consortium .World J MicrobiolBiotechnol.,2008;24:55-60.DOI:10.1007/s11274-007-9437-0.
CrossRef - Lily, M. K., Bahuguna, A., Dangwal, K.,Garg, V. Degradation of Benzo [a] Pyrene by a novel strain Bacillus subtilis BMT4i (MTCC 9447). EnvMicrobiol.,2009;40:884-892. DOI:10.1590/S1517-83822009000400020.
CrossRef - Christova, N., Tuleva, B.,Nikolova-Damyanova, B. Enhanced Hydrocarbon Biodegradation by a Newly Isolated Bacillus subtilis Strain. Z. Naturforsch., 2004;59:205-208.
CrossRef - Jalilzadeh Yengejeh, R., Sekhavatjou, M.S., Maktabi, P., Arbab Soleimani, N., Khadivi, S.,Pourjafarian, V. The Biodegradation of Crude Oil by Bacillussubtilis Isolated from Contaminated Soil in Hot Weather Areas. Int. J.Environ. Res.,2014;8: 509-514.
- Kafilzadeh, F.,Hoshyari Pour, F. Degradation of naphthalene, phenanthrene and pyrene by Pseudomonas sp and Corynebacterium sp in the landfills.Int. J. Biosci., 2012;2:77-84.
- Fava, F., Armenante, P., Kafkewitz, D. Aerobic degradation and dechlorination of 2-chlorophenol, 3-chlorophenol and 4-chlorophenol by a Psedomonas pickettii srain. Lett ApplBiotechnol.,1995;43: 171-177.
- Lin, C., Gan, L.,Chen, Z-L. Biodegradation of naphthalene by strain Bacillus fusiformis (BFN). J Hazard Mater., 2010; 182: 771-777. DOI:10.1016/j.jhazmat.2010.06.101.
CrossRef - Das, K.,Mukherjee, A.K. Crude petroleum-oil biodegradation eYciency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. BioresourTechnol., 2007;98:1339–1345. DOI:10..1016/j.biortech.2006.05.032.
CrossRef - Mutalik, S. R ., Vaidya, B. K., Joshi, R. M. , Desai,K.,Mene, S. N. Use of response surface optimization for the production of biosurfactant from Rhodococcus spp. MTCC 2574. BioresourceTechnol.,2008;99:7875-7880. DOI:10.1016/j.biortech.2008.02.027.
CrossRef - Prassana, N., Saravanan, N., Geetha, P., Shanmugaprakash, M.,Rajasekaran, P. Biodegradation of phenol and Toluene by Bacillus sp., Psedomonas sp., and Staphylococcus sp., Isolated from pharmaceutical Industrial Effluent. AdvancedBioTech., 2008; 20-24.
- Abdel-Mawgoud, A.M., Aboulwafa, M. M., Haleem, Abdel.,Hassouna N. Optimization of surfactin production by B.subtilis Isolate BS5. ApplBiochemBiotechnol., 2008; 150:305-325. DOI:10.1007/s12010-008-8155-x.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





