How to Cite | Publication History | PlumX Article Matrix
Asep A. Prihanto1, Abdul A. Jaziri1 and Ima Y. Perwira2
1Department of Fishery and Marine Science, Faculty of Fisheries and Marine Science, Brawijaya University, Veteran street, Malang, 651245, East Java Indonesia.
2Department of Aquatic Resources Management, Faculty of Marine Science and Fisheries, Udayana University, Bali - Indonesia.
Corresponding Author E-mail: asep_awa@ub.ac.id
DOI : http://dx.doi.org/10.13005/bbra/2283
ABSTRACT: Neutral protease producing bacterium Bacillus substilis UBT7 strain isolated from Terasi, an Indonesian fermented fish grew in shake flask and fermenter, and produced protease at 37 °C, pH 7 for 24 hour. After three purification steps, the enzyme was successfully purified. This neutral protease had an apparent molecular mass of 32 kDa as estimated by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis. The enzyme was active optimum on 50 °C. This indicated that the enzyme was slightly thermozyme. The stability was decreased on 40 °C. The enzyme was optimum in pH 7. The enzyme was somehow enhanced with Fe2+ and K2+ metal ionsbut not by the addition of Ni2+.
KEYWORDS: Terasi; Bacillus substilis UBT7; neutral protease; fermented fish; Indonesia
Download this article as:| Copy the following to cite this article: Prihanto A. A, Jaziri A. A, Perwira I. Y. Purification and Characterization of Neutral Protease from Bacillus Substilis Ubt7 Isolated from Terasi, Indonesian Fermented Fish. Biosci Biotech Res Asia 2016;13(3). |
| Copy the following to cite this URL: Prihanto A. A, Jaziri A. A, Perwira I. Y. Purification and Characterization of Neutral Protease from Bacillus Substilis UBT7 Isolated from Terasi, Indonesian Fermented Fish. Biosci Biotech Res Asia 2016;13(3). Available from: https://www.biotech-asia.org/?p=15709 |
Introduction
Terasi is one of fermented fish or shrimp products from Indonesia. It is a traditional condiment commonly used as a flavor enhancer in every Indonesian household. Terasi is made by mixing fish or shrimp with salt and drying it under the sun. Fermentation process usually takes 6-8 months until the desired terasi aroma has been developed. Terasi contains free glutamic acid, and it is rich in umami taste (Hajeb and Jinab, 2012).
The most important process which associated with Terasiprocessing is amino acid degradation by proteases (Prihantoet al., 2013). Protease hydrolyzes long protein chains into smaller fragments by cleaving the peptide bonds. Proteases have been used commercially in various industries such as food, pharmaceutical, detergent, photographic, and biotechnological industries (Gupta et al., 2002). They make up about 60% of the total of global sale of enzymes, which are predicted to exceed $2.9 billion (Sarrouhet al., 2012). The sources of protease consist of plants, animals and microorganisms. Moreover, microbial protease is preferred than plant or animal sources due to broad biochemical diversity, rapid growth, limited space required for cell cultivation and ease to be produced (Nigam et al., 2012).
In the present research, various bacterial strains have been isolated and screened for extracellular protease production from fermented fish or shrimp paste (Prihantoet al., 2013). However, bacterial strain identity confirmed by 16S rDNA is still poorly investigated, as well as its protease enzyme has not been purified and characterized yet. In this study, we attempted to identify bacterial strain by conducting 16S rDNA analysis and purify the protease enzyme from Bacillus substilisisolated from Terasi using anion exchange chromatography coupled with gel filtration.
Materials and Methods
Bacterial strain and media
A number of bacterial strains were isolated from Terasi. 1 gram of Terasi was mixed with 9 ml normal saline. Then it was serially diluted from 10−1 to 10−6 ratio with normal saline, and 100 μl of each diluted samples was inoculated on Luria–Bertani’s (LB) agar medium. The plates were incubated at 37 °C for 24 h. The isolated colonies were selected to obtain pure bacterial strains. Pure bacterial isolates were grown at 37 °C for 24 h onto the LB agar medium containing 1.0% skim milk (SM) for the screening of proteolysis activity. The clear zone of SM hydrolysis around the colony confirmed the protease production. The maximum protease producing bacterial strain was selected and identified on the basis of morphological and biochemical characteristics.
Identification of strain UB-7
StrainUB-7 was identified by morphological and biochemical tests and its 16S ribosomal DNA (16S rDNA) sequence (Sneathet al., 1986). The DNA extraction procedure for the sequencing of 16S rDNA from bacterial cells and sequence data analysis were described previously (Maeda and Suga, 1992). The 16S rDNA of strain UB-7 was amplified by polymerase chain reaction (PCR) using primers 20F (5′-GTAATCGTCGGCCAGTA GAGTTTGATCCTGGCTC-3′) and 1510R (5′-CAGGAA ACAGCTATGACCGGCTACC TTGTTACGACT-3′). PCR amplification was carried out for 30 cycles of 94°C for 2 min, 98°C for 10 s, 61°C for 30 s and 68°C for 1 min. A PCR product purification kit (The Wizard® SV Gel and PCR Clean-Up System; Promega, USA) was used for purification of the amplified DNA. The 16S rDNA sequences were determined with a ABI 3130xl DNA Sequencer (Applied Biosystems, USA) using sequencing primers, F1 (5′-GAGTTTGATCCTGGCT CAG-3′); F3 (5′-GTCCCGCAACGAGCGCAAC-3′); R1 (5′-GTATTACCGCGGCTGCTGCTG G-3′); R2 (5′-CATCGTTTACGGCGTGGAC-3′); R3 (5′-TTGCGCTCGTCGTTGCG GACT-3′); and R4 (5′-ACGGGCGGTGTGATACAAG-3′), and the sequences were compared with the 16S rDNA sequences in the GenBank database using BLAST. Phylogenetic tree analysis followed Dereeperet al., (2008) method.
Protease assay
The protease activity assay was conducted through the Anson method (Anson, 1938) with some modifications. The enzyme solution (0.5 ml) was incubated with 5 ml of 1% (w/v) casein solution prepared in Tris–HCl buffer (pH 8.0) for 20 min at 80 °C. The reaction was stopped by addition of 1.5 ml of 0.3 M TCA (Tricarboxylic acid). The contents were centrifuged at 10,000 rpm for 10 min at 4 °C and the amino acids produced by the degradation of casein were measured as tyrosine through the method of Folin and Ciocalteu. One unit of protease was defined as “the amount of enzyme that hydrolysed casein to produce colour equivalent to 1.0 μM of tyrosine per minute under optimized reaction conditions”.
Protein determination
The total protein was determined through Lowry method with bovine serum albumin as the standard (Lowry et al., 1951). Absorbance at 280 nm was used for monitoring protein in column elutes (Warburg and Christian, 1942).
Purification of protease
A crude protease solution was obtained by centrifugation of the culture broth at 10000 rpm for 10 min. The cell free supernatant precipitated with 60% saturation of ammonium sulfate and incubated overnight at 4 °C. The precipitated protein was obtained by centrifugation for 20 min at 7000 rpm at 4 °C. The obtained pellet was re-suspended in a 10 mL of ice-cold 0.2 M phosphate buffer pH, 8 and then subjected to a process of dialysis against the same buffer to get rid of the excess of ammonium sulfate. The dialyzed protease solution was purified with an AKTA explorer chromatography system (GE ÄKTA Explorer, USA). First, the protease solution was applied to an anion-exchange HiTrap Q HP column (5 ml) (GE Healthcare) that had been pre-equilibrated using buffer A. The protease was eluted with a linear gradient of 0 to 1.0 M NaCl, and the peak fractions were collected. The fractions were further applied to a Superdex 200 10/300 GL gel filtration column (GE Healthcare) pre-equilibrated with buffer A containing of 0.15 M NaCl. The single fraction of neutral protease was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein concentration was determined the by Bradford method with bovine serum albumin as the standard.
Characterization of protease
Effect of temperature
The optimum temperature for maximum catalytic activity of protease was determined by measuring enzyme activity in different incubation temperatures ranging from 30 °C to 70 °C for 30 minute.
Thermal stability
The thermal stability of protease was measured by pre-incubation of enzyme solution at 30 – 70 °C for ten minutes, after definite time, the residual activity ofthe enzyme were assayed.
Effect of pH
The optimum pH for protease activity were investigated. Various pH levels ranging from pH 4 to pH 12 at constant substrate concentration and temperature were used for determining optimum pH.
pH stability
The pH stability of protease was measured by pre-incubation of enzyme solution at pH 4 to pH 12 for ten minutes. After definite time interval the enzyme were assayed for its residual activity.
Effect of metal ions
For investigating the effect of metal ions and potential inhibitor of B. substilisneutral protease,the enzyme were reacted in the present of these substances on the concentration of 5 mM. The activity were examined under standard assay.
Result and Discussion
Strain identification
Isolated strain was identified by phylogenetic analysis based on 16S rDNA sequence. The sequence was deposited on NCBI (accession number,KR780584). This sequence has a 99.0, and 99.3 similarities with those deposited sequences namely Bacillus subtilisstrain setapak 8, andBacillus substilis, respectively. Furthermore, SEM analysis revealed that the strain was rod bacterium with 1.5~2.5 µm size(unpublished data). Taxonomic characteristics in order to analysis of strain was also performed by API system. The isolate is gram positive, catalase-positive, oxidase-positives, gelatinase-positives and NO3 negatives. All of these result indicated that the strain was Bacillus sp. The phylogenetic tree analysis showed that this isolate was newly strain of Bacillus substilis (Fig. 1). Hence we called this isolate Bacillus substilis UBT7.
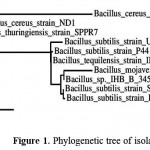 |
Figure 1: Phylogenetic tree of isolated bacterium.
|
Production, Purification and Characteristics of Enzyme
Bacillus substilisUBT7 was incubated in LB medium with shaking orbital (110 rpm) for 3 days. The optimum production of enzymes was reached after 34 hours (Fig 2). The enzyme was pure after three steps of purifications. An apparent molecular mass of 32 kDa was found.
The summary of purification steps were shown in Table 1.
Table 1: Summary of purification of protease from B. substilis UBT7
| Purification step | Total activity (U) | Total protein (mg) | Specific activity (U/mg) | Purification (fold) | Yield
(%) |
| Cell-free extract | 840 | 310 | 2.71 | 1.0 | 100 |
| Precipitation 60% | 320 | 191 | 1.68 | 0.6 | 38 |
| HiTrap Q HP | 160 | 32 | 5.00 | 1.8 | 19 |
| Superdex 200G | 39 | 4.3 | 9.07 | 3.3 | 4.6 |
Forty three fold of purification was achieved in these purification step. Specific activity of the enzyme was increase about three times in the final step of the purification. The final purification step resulted in 3.3 fold purification with the yield of 4.6%.
 |
Figure 2: SDS-PAGE analysis of purified B. cereusneutral protease. Lane 1 molecular mass markers, lane 2 precipitation step, lane 3 Hitrap Q HP, lane 4 Superdex 200G.
|
The optimum temperature of the enzyme was 50 ºC and the enzyme activity started to decrease on 40 ºC (Fig 3). The enzyme can be categorized as thermo enzyme. The enzyme which has an optimum activity above 40 is categorized as thermophile enzyme. Thermoenzyme neutral protease was also reported by Aqelet al., (2012). This enzyme even stable at 100 ºC
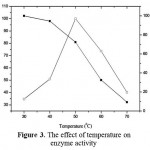 |
Figure 3: The effect of temperature on enzyme activity.
|
The pH 7 is the optimum pH for activity of neutral protease purified from B. substilis UB7. The enzyme also stable on pH 6-8 (Fig. 4). This indicated that the enzyme is neutral protease. Neutral protease also isolated from Bacillus substilisisolated from gut of Bombimorix (Wang et al., 2016).
 |
Figure 4: The effect of pH on enzyme activity.
|
The protease activity was enhanced with addition of 5mM FeCl2 andKCl, resulting in the relative activityof 111.19 and 142%, respectively (Tab 2.). In contrast, the protease was severely inhibited upon the addition of 5 mM NiSO4 resulting in relative activity of 80, 5 %, respectively. Protease required a divalent cation like Ca2+, Fe2+, Na2+ and K2+ or combination of these cations for its maximum activity (Kumar et al., 1999).
Tabel 2: The effect of metal ions on enzyme activity
| Metal ion | Relative activity (%) |
| None | 100 |
| Ca⁺2 | 103.5 |
| Ni | 80.5 |
| Zn | 100.33 |
| Mg | 100 |
| Mn | 97..2 |
| Fe | 111.9 |
| Na | 103.2 |
| Co | 96.8 |
| K | 142.9 |
It is also suggested that most proteases enzyme requires Fe2+ and Ca2+ to facilitate their optimum activity (Takamiet al., 1989). It was found that Fe2+, Ca2+ increased neutral protease activity. Neutral protease derived from Pseudomonas aeruginosa indicated the similar trend. The enzyme needs Fe2+, Ca2+,Na2+ for their optimum activity (Lu and Chang, 1996). It seems that Fe2+ is common enhancer for neutral protease (Wang et al., 2013)
In conclusion, B. substilisUBT7 have been isolated from Terasi, an Indonesia Fermented fish. The protease was produced on LB medium. It is 32 kDa of molecular weight. The character of the enzyme indicated that the optimum temperature, pH, are, 50 ºC, 7, respectively. With the present of Fe2+ and K2+, the enzyme activity are enhanced. Further studies are necessary to find out the applications of studied enzyme.
References
- Anson, M.L.The estimation of pepsin, trypsin, papain and cathepsin with hemoglobin. J. Gen. Physiol1938; 22:79-89.
CrossRef - Aqel H.,vAl-Quadan F., Yousef TK. A novel neutral protease from thermophilicBacillus strain HUTBS62. J. BioSci. Biotech. 2012; 1: 117-123.
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist.Nucleic Acids Res2008; 36: W465–W469
CrossRef - Gupta R, Beg QK, Lorenz P. Bacterial alkaline proteases: Molecular approaches and industrial application. Appl. Microbiol. Biotechnol2002; 59: 15-32
CrossRef - Hajeb P and Jinap S. Fermented Shrimp Products as Source of Umami in Southeast Asia. J Nutr Food Sci2012;S10:006
- Kumar CG, Tiwari MP, Jany KD. Novel alkaline serine proteases from alkalophilic Bacillus spp.: purification and some properties. Process Biochem1999; 34: 441-449
CrossRef - Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature1970. 227:680-685.
CrossRef - Lowry O.H, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem 1951;193: 265-275
- Lu, S F., and Chang PP. A thermostable neutral protease from Pseudomonas aeruginosa CCRC 15541 S. Letters in Applied Microbiology 1996;22: 5-9
CrossRef - Nigam P, Singhal,V.K, Vidyarthi AS. Studies on production, characterization and applications of microbial alkaline proteases. Int. J. Advanc. Biotechnol. Res. 2012; 3: 653- 669
- Prihanto AA., Darius.,Firdaus,. Proteolyticand Fibrinolytic Activities OfHalophilic Lactic Acid Bacteria From Two Indonesian Fermented Foods. Journal of Microbiology, Biotechnology and Food Sciences2013; 2: 2291-2293
- Sarrouh B, Santos TM, Miyoshi A, Dias R, Azevedo V. Up-To- Date Insight on Industrial Enzymes Applications and Global Market. J Bioprocess Biotechniq2012;S4:002. doi:10.4172/2155-9821.S4-002
CrossRef - Sneath, P. H. A., Mair, N. S., Sharpe, M. E. & Holt, J. G. (editors). Bergey’s Manual of Systematic Bacteriology, vol. 2. Baltimore: Williams & Wilkins1986.
- Wang, J., Xu, A., Wan, Y., Li, Q. Purification and Characterization of a New Metallo-Neutral Protease for Beer Brewing from Bacillus amyloliquefaciens SYB-001. ApplBiochemBiotechnol2013; 170: 2021–2033
CrossRef - Wang, Z. Yang, W., Sun, L., Zhu, B., Li D., Liu C. Characterization of a Neutral Protease Gene of Bacillus subtilis Isolated from the Guts of Bombyxmori. Pakistan J. Zool2016;48: 179-185
- Warburg, O. and W. Christian.Isolation and crystallization of enolase. Biochem. Z 1942;310:384-421

This work is licensed under a Creative Commons Attribution 4.0 International License.





