How to Cite | Publication History | PlumX Article Matrix
Soodabeh khalili¹,2, Mahmoud Shekari Khaniani2, Naser Aghamohammdzade3, Abolfazl Akbarzadeh4 and Sima Mansoori Derakhshan¹,2*
1Neurosciences Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
2Department of Medical Genetics, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran.
3Department of Internal Medicine, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran.
4Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
DOI : http://dx.doi.org/10.13005/bbra/2335
ABSTRACT: Nesfatin-1, which is derived from the NEFA/nucleobindin 2 (NUCB2) precursor, is a satiety factor secreted by several tissues, including the hypothalamus. NUCB2 gene expression positively correlates with insulin secretory capacity, as well as with insulin and glucagon gene expression in human islets. Therefore, we hypothesized that polymorphisms in the NUCB2 gene promoter influence the susceptibility for the development of diabetes. In the current study, we investigated the association of NUCB2 polymorphisms in the promoter region and 5′ UTR, exon1, and 5′ part of intron 1 of NUCB2 gene with type II diabetes. A total of 200 patients with type II diabetes and 200 healthy controls subjects were enrolled in this study. The 5′ part of NUCB2 variants was assessed using PCR and direct sequencing. Twelve variants, including rs186174(c.-427A > C), rs214088 (c.-406C > G), rs4757506(c.254A > G), rs369209853 (c.- 205G > A), rs214087 (c.-166G > C), rs373592192 (c.-281G > A), rs182903196 (c.-308G > A), rs 370538176 (c.-426C > G), rs 190662423 (c.-612C > A), rs115148100 (c.-653G > A), rs374389403 (c -233T > c), and rs377756452 (c -207G > A), were identified. Analysis of the results showed that the frequency of five of the polymorphisms [rs 214088 (c.-406C >G), rs 4757506 (c.254A > G), rs369209853 (c.-205G > A), rs214087 (c.-166G > C), and rs373592192 (c.-281G > A)] is significantly different between patients and controls subjects (P< 0.05), which indicates the association of these polymorphisms with type 2 diabetes. Although the relationship between the promoter region of NUCB2 gene and T2D remains uncertain at present, These findings may support the role of NUCB2 gene in the regulation of the regulation of blood glucose and the increased type 2 diabetes in humans.
KEYWORDS: NUCB2; type 2 diabetes; Single nucleotide polymorphism
Download this article as:| Copy the following to cite this article: Khalili S, Khaniani M. S, Aghamohammdzade N, Akbarzadeh A, Derakhshan S. M. The Association of Nucleobindin 2 Gene (NUCB2) Variants with Type 2 Diabetes Mellitus Among Iranian Azeri-Turkish Population. Biosci Biotech Res Asia 2016;13(3) |
| Copy the following to cite this URL: Khalili S, Khaniani M. S, Aghamohammdzade N, Akbarzadeh A, Derakhshan S. M. The Association of Nucleobindin 2 Gene (NUCB2) Variants with Type 2 Diabetes Mellitus Among Iranian Azeri-Turkish Population. Biosci Biotech Res Asia 2016;13(3).Available from: https://www.biotech-asia.org/?p=15209 |
Introduction
NUCB2/nesfatin and its proteolytically cleaved product nesfatin-1 [1] were originally identified as proteins secreted by the hypothalamus and involved in feeding regulation. NUCB2 consists of 14 exons, spanning 54785 nucleotides (accession number NC_000011.9). It codes for a precursor protein with a signal peptide containing 24 amino acids and a protein structure containing 396 amino acids [2] and is highly conserved among rodents and humans as well as within nonmammalian vertebrate species, indicating its importance [3]. It is post-translationally divided into three segments by the prohormone convertase (PC), which includes the N-terminal segment NUCB2/nesfatin-1 (1–86), two C-terminal peptide nesfatin-2 (85–163), and nesfatin-3 (166–396) [4]. Nesfatin-1 is an anorexigenic and insulinotropic peptide found abundantly in the hypothalamus, pancreas, and gastric oxyntic mucosa [4]. It possesses anti-hyperglycemic properties, but these are likely to be insulin dependent [4]; therefore , the dysfunction of nesfatin-1 could be implicated in metabolic disorders, particularly type 2 diabetes (T2D), and consequently, it could act as an anti-diabetic factor by enhancing both insulin secretion and insulin action [5].
A major function of nesfatin-1 is the inhibition of food intake and regulation of blood glucose in time , dose , and insulin-dependent manners [6], and it also may have an important role in insulin resistance of T2D [7]. The relationship between plasma levels of nesfatin-1 and the presence of T2D correlates either negatively or positively [8, 9]. The reasons for this discrepancy remain unclear, but it may be derived from the differences in the severity of disease; different patient characteristics, for example, body mass index; different assessment methods (ELISA recognizing NUCB2 and nesfatin-1 vs. sandwich-type ELISA recognizing only nesfatin-1); and gender differences (females vs. males) [2, 10]. However, the roles of nesfatin-1 in the pathogenesis of insulin resistance and T2D are presently not well understood [11].
As far as NUCB2 gene is concerned, the promoter of this gene has not yet been investigated. Therefore, we intended to investigate the known single nucleotide polymorphisms (SNPs) in the promoter region of this gene in T2D patients. On the other hand, the gene located on the short arm of chromosome 11 is associated with diabetes, and hence it is important to evaluate the promoter region. In the present study, we genotyped 12 SNPs in the NUCB2 gene in Azeri-Turkish population of Iran .The genotypes and alleles distributions of these 12 SNPs are shown in Table 1 and assessed the association between NUCB2 and T2D patients and control subjects.
Methods and Materials
Subjects and Sample Collection
This case-control study was approved by the research ethics committee of Tabriz University of Medical Sciences, and each patient or control individual enrolled in this study gave a written informed consent and the approval form for genetic analysis. The study population consisted of 200 normoglycemic control subjects (105 men and 95 women; aged 47.30 ± 2.29 years; body mass index (BMI) 24.96 ± 0.92 kg/m2) and 200 T2D patients (111 men and 89 women; aged 53.55 ± 1.80 years; body mass index (BMI) 26.46 ± 0.46 kg/m2) from the outpatient Endocrinology clinic of Imam Reza university hospital, Tabriz, Iran. Table 2 shows the diabetes-related clinical and biochemical parameters of all the study subjects.
The control subjects consisted of healthy, unrelated, non-diabetic spouses of the T2D patients. None of them had a family history of diabetes or were taking any medication known to change glucose tolerance. Patients with type 1 diabetes mellitus, hypertension, heart failure, pregnancy, acute or chronic infectious disease, liver or kidney disease, and cancer were excluded from the study.
The T2D patients were recruited after a detailed investigation of their medical records for symptoms, use of an oral hypoglycemic agent and insulin, measurement of fasting blood glucose, and other related anthropometric parameters.
Primers were designed using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/ primer3_www.cgi) and the NUCB2 genomic sequence, with the accession number NG_003180 [Homo sapiens cytochrome P450, family 2, subfamily D, polypeptide 6 genomic region (C YP2D6 on chromosome 22)].
There are 12 SNPs for the human NUCB2 gene listed in the SNP database of the National Center for Biotechnology Information [12]; therefore, we screened 700 bp of the NUCB2 gene in T2D patients and identified the 12 SNPs. To determine the SNPs, the DNA sequences of the variants were retrieved from the dbSNP database [12].The genotype and allele frequencies of all the SNPs were identified and determined in all the study participants. Data analysis was performed using the Statistical Package for Social Sciences-SPSS computer software for Windows (version 21.0).
DNA Analysis
Genomic DNA was extracted from peripheral blood lymphocytes using the salting-out method, as previously described. According to the NUCB2 sequence, one pair of primers (F: 5′–GTGGGTGCGGTTACAAATG–3′ and 5′ TCTGCTCCACTCGCAGCT 3′) was designed to amplify the 700 bp fragment consisting of the promoter, the exon 1, and the 5 ́ end of the first intervening sequence of NUCB2 gene. Polymerase chain reaction (PCR) was performed with a final reaction volume of 25 μl, which included 100 ng of genomic DNA, 10 pM of each primer, 1.5 U of Taq DNA polymerase, 2.5 μl of the 10 × buffer, 1.5 mM of MgCl2, and 0.25 mM of dNTPs. The reaction was performed on a thermal cycler (Bio-Rad thermal cycler, My Cycler TM thermal cycler, USA), with initial denaturation at 94°C for 5 min, followed by 30 cycles at 94°C for 30 s, 62°C for 1 min, and 72°C for 30 s, and a final extension at 72°C for 5 min. The PCR products were analyzed by sequencing on an ABI 3700 automatic sequencer machine (Macrogen, South Korea).
Statistical Analysis
Data were analyzed using the general linear model by investigating the association between the variants and T2D. The differences between the genotype distribution and allele frequency in the study groups were analyzed using chi-square test. The odds ratios (OR) for each section were compared through the use of a fixed effects model. Statistical heterogeneity was evaluated using χ² to assess the appropriateness of pooling the individual study results.
Results and Discussions
Results
To determine the sequence variation in the 5′ region of nucb2 gene, we examined a 0.7 kb fragment comprising the promoter region, exon 1, and first intron. Sequencing this region revealed 12 SNPs in all the 400 tested subjects, which included 7 SNPs in the promoter regions, 4 SNPs in exon 1, and 1 SNP in the first intron (Figure 1).
All variants were in agreement with the Hardy–Weinberg equilibrium. In total, 7 SNPs were observed in both T2D patients and healthy control subjects, while 5 SNPs showed a significant difference in the association between these two groups (Table 3).
Our results showed that rs 214088 (c.-406C >G), rs 4757506 (c.254A > G), rs369209853 (c.-205G > A), rs214087 (c.-166G > C), and rs373592192 (c.-281G > A) variants are associated with T2D. The genotype and allele distributions of rs214088 (c.-406C > G) differed significantly between the T2D patients and control subjects (P < 0.001). The C/C homozygote and C/G heterozygote genotypes of this variant were more frequent in the T2D group, while the G/G homozygote genotype was found more frequently in the control group. The G/G genotype and the G allele were more common in control than in T2D subjects. These results confirm that rs 214088 variant has the strongest association with T2D (p = 0.000).
The genotype of rs 4757506 (c.254A > G) differed significantly between the T2D patients and the control subjects (both P < 0.001). A comparison of the frequency of A/A genotypes in the rs 4757506 variant between the two groups showed 24.5% higher occurrences of rs 4757506 in control than in T2D subjects (53.0% vs.77.5%), while the A/G genotype was found more frequently in the T2D patients (39.0% in T2D) and the G/G genotype showed the same frequency (8.0%) in both the study groups.
The genotype and allele distributions of rs214087 (c.-166G > C) differed significantly between the T2D patients and control subjects (both P < 0.001), and the C/C and C/G genotypes were more frequent in T2D patients, while the G/G genotype was found more frequently in the control group. The genotype and allele distributions of rs369209853 (c.-205G > A) and rs373592192 (c.-281G > A) differed significantly between the two study groups (both P < 0.001), with an evident linkage to diabetes.
However, the genotype and allele distributions of the other seven SNPs were not different between the T2D patients and control subjects. The SNPs including rs115148100 (c.-653G > A), rs190662423 (c.-612C > A), rs186174 (c.-427A > C), rs 370538176 (c.-426C > G), rs182903196 (c.-308G > A), rs374389403 (c -233T > c and rs377756452 (c -207G > A) did not show any significant association with diabetes (p> 0.05). The lowest correlation was observed for rs377756452 (c -207G > A), which showed an almost similar frequency in the two study groups. The remaining SNPs did not show any linkage with diabetes, and they were found in similar frequencies in both the groups.
Table 1: Characteristics of study population
| Controls (200) | Patients (200) | |
| Sex (M/F) | F:95
M: 105 |
F:89
M:111 |
| Age (years) | 47.30 ± 2.29 | 53.55 ± 1.80 |
| Body mass index (kg/m²) | 24.96 ± 0.92 | 26.46 ± 0.46 |
| HbA1C (%) | 5.4 ± 0.3 | 7.6 ± 0.29 |
| Plasma glucose (mmol/L) | 5.33 ± 0.26 | 8.41 ± 0.36 |
| Total cholesterol (mmol/L) | 5.2 ± 0.9 | 5.4 ± 0.32 |
| Triglyceride (mmol/L) | 1.2 ± 0.2 | 1.4 ± 0.8 |
| Systolic blood pressure (mmHg) | 128 ± 15 | 134 ± 18 |
| Duration of T2D | – | 6.44 ± 0.91 |
| Treatment
Diet only (%) Oral agents (%) Insulin (%) |
– |
7.7 66.7 25.6 |
Table 2: List of Single nucleotide polymorphism identified in this study
| Polymorphism | Nucleotide position | Substitution | Location |
| rs115148100 (c.-653G > A) | -653 | G → A | Intronic |
| rs190662423 (c.-612C > A) | -612 | C → A | Intronic |
| rs186174 (c.-427A > C) | -427 | A → C | Intronic |
| rs370538176 (c.-426C > G) | -426 | C → G | Intronic |
| rs214088 (c.-406C > G) | -406 | C → G | Intronic |
| rs182903196 (c.-308G > A) | -308 | G → A | Intronic |
| rs4757506c.254A > G | 254 | A → G | Intronic |
| rs374389403 (c -233T > C) | -233 | T → C | 3´UTR/5´UTR |
| rs377756452 (c -207G > A) | -207 | G → A | 3´UTR/5´UTR |
| rs369209853 (c.-205G > A) | -205 | G → A | 3´UTR/5´UTR |
| rs214087 (c.-166G > C) | -166 | G → C | 3´UTR/5´UTR |
| rs373592192 (c.-281G > A) | -281 | G → A | Intronic |
Discussions
As nesfatin-1 has been suggested to have insulin-dependent anti-hyperglycemic properties [4], dysfunction of nesfatin-1 could be implicated in T2D. To the best of our knowledge, no genetic research has been performed to investigate the association of NUCB2 genetic variants with the development of T2D. We performed sequencing analyses to assess the differential variants of NUCB2 promoter, exon 1, and the 5′ end of the first intervening sequence of NUCB2 gene in T2D patients and normal controls subjects.
Table 3: Allele distributions of NUCB2 gene in T2D patients and control subjects.
| Genotype | Cases | Controls | P Value | OR (CI 95%) | |||
| Frequency | (%) | Frequency | (%) | ||||
| rs115148100 | AA | 22 | 5.5% | 27 | 6.8% | 0.341
|
– |
| AG | 14 | 3.5% | 8 | 2.0% | |||
| GG | 164 | 41.0% | 165 | 41.3% | |||
| Allelic frequency | G | 342 | 42.8% | 338 | 42.3% | 0.692 | 1.081 (0.734–1.595) |
| A | 58 | 7.2% | 62 | 7.8% | |||
| rs190662423 | AA | 5 | 1.3% | 6 | 1.5% | 0.256 | – |
| CA | 10 | 2.5% | 4 | 1.0% | |||
| CC | 185 | 46.3% | 190 | 47.5% | |||
| Allelic frequency | C | 380 | 47.5% | 384 | 48.0% | 0.495 | 0.792 (0.404–1.551) |
| A | 20 | 2.5% | 16 | 2.0% | |||
| rs186174 | AA | 0 | 0.0% | 0 | 0.0% | 0.001 | – |
| CA | 35 | 8.8% | 14 | 3.5% | |||
| CC | 165 | 41.3% | 186 | 46.5% | |||
| Allelic frequency | A | 35 | 4.4% | 14 | 1.8% | 0.002 | 2.644 (1.4–4.994) |
| C | 365 | 45.6% | 386 | 48.3% | |||
| rs370538176 | CC | 182 | 45.5% | 184 | 46.0% | 0.225 | – |
| CG | 14 | 3.5% | 8 | 2.0% | |||
| GG | 4 | 1.0% | 8 | 2.0% | |||
| Allelic frequency | G | 22 | 2.8% | 24 | 3.0% | 0.761 | 0.912 (0.502–1.655) |
| C | 378 | 47.3% | 376 | 47.0% | |||
| rs214088 | CC | 23 | 5.8% | 19 | 4.8% | 0.0001 | – |
| CG | 98 | 24.5% | 47 | 11.8% | |||
| GG | 79 | 19.8% | 134 | 33.5% | |||
| Allelic frequency | C | 256 | 32.0% | 315 | 39.4% | 0.0001 | 0.480 (0.350–0.657) |
| G | 144 | 18.0% | 85 | 10.6% | |||
| rs182903196 | AA | 14 | 3.5% | 7 | 1.8% | 0.013 | – |
| AG | 7 | 1.8% | 20 | 5.0% | |||
| GG | 179 | 44.8% | 173 | 43.3% | |||
| Allelic frequency | G | 365 | 45.6% | 366 | 45.8% | 0.9 | 0.969 (0.591 -1.587) |
| A | 35 | 4.4% | 34 | 4.3% | |||
| rs4757506 | AA | 102 | 25.5% | 146 | 36.5% | 0.0001 | – |
| AG | 75 | 18.8% | 29 | 7.2% | |||
| GG | 23 | 5.8% | 25 | 6.3% | |||
| Allelic frequency | G | 121 | 15.1% | 79 | 9.9% | 0.001 | 1.762 (1.272–2.441) |
| A | 279 | 34.9% | 321 | 40.1% | |||
| rs374389403 | CC | 8 | 2.0% | 17 | 4.3% | 0.128 | – |
| TC | 12 | 3.0% | 8 | 2.0% | |||
| TT | 180 | 45.0% | 175 | 43.8% | |||
| Allelic frequency | C | 28 | 3.5% | 42 | 5.3% | 0.08 | 0.642 (0.389–1.057) |
| T | 372 | 46.5% | 358 | 44.8% | |||
| rs377756452 | AA | 4 | 1.0% | 6 | 1.5% | 0.638 | – |
| AG | 5 | 1.3% | 3 | 0.8% | |||
| GG | 191 | 47.8% | 191 | 47.8% | |||
| Allelic frequency | G | 387 | 48.4% | 385 | 48.1% | 0.7 | 1.16 (0.545–2.47) |
| A | 13 | 1.6% | 15 | 1.9% | |||
| rs369209853 | AA | 6 | 1.5% | 28 | 7.0% | 0.0001 | – |
| AG | 21 | 5.3% | 9 | 2.3% | |||
| GG | 173 | 43.3% | 163 | 40.8% | |||
| Allelic frequency | G | 367 | 45.9% | 335 | 41.9% | 0.001 | 2.158 (1.365–3.365) |
| A | 33 | 4.1% | 65 | 8.1% | |||
| rs214087
|
CC | 37 | 9.3% | 33 | 8.3% | 0.0001 | – |
| CG | 105 | 26.3% | 63 | 15.8% | |||
| GG | 58 | 14.5% | 104 | 26.0% | |||
| Allelic frequency | G | 221 | 27.6% | 272 | 34.0% | 0.0001 | 0.581 (0.436–0.775) |
| C | 179 | 22.4% | 128 | 16.0% | |||
| rs373592192
|
AA | 5 | 1.3% | 25 | 6.3% | 0.0001 | – |
| AG | 19 | 4.8% | 3 | 0.8% | |||
| GG | 176 | 44.0% | 172 | 43.0% | |||
| Allelic frequency | G | 371 | 46.4% | 347 | 43.4% | 0.005 | 1.954 (1.214–3.145) |
| A | 29 | 3.6% | 53 | 6.60% | |||
T2D is a genetically heterogeneous disorder and is associated with insulin resistance and impaired insulin secretion. The common forms of T2D are believed to be complex polygenic diseases [13]. Thus far, genetic association studies involving NUCB2 polymorphisms have been conducted in relation to obesity and metabolic syndrome. In 2011, Zegers et al. reported that three single nucleotide polymorphisms were associated with obesity in male Caucasian subjects [14]. In another cohort of 471 obese children, seven sequence variants of NUCB2 gene were detected [15]. However, these variants did not result in differences in the expression of NUCB2 and plasma nesfatin-1 levels from those observed in body weight matched controls subjects [16].
NUCB2 gene expression correlated positively with insulin secretory capacity, as well as with insulin and glucagon gene expression in human islets [17].
Zhang et al. reported a positive relationship between plasma nesfatin-1 levels and impaired glucose tolerance. In contrast, Li et al. reported that fasting plasma nesfatin-1 levels are lower in T2D mellitus than in normal controls subjects .The reasons for this discrepant results are still unclear; however it was suggested that the difference is due to the differences in disease severity or the different patient characteristics, such as body mass index [18].
The relationship between plasma levels of nesfatin-1 and the presence of T2D mellitus have been reported to correlate either negatively or positively [18] and associated with BMI, plasma insulin [19].
The chi-square analysis revealed significant polymorphisms of the rs214088 (c.-406C > G), rs4757506 (c.254A > G), rs369209853 (c.-205G > A), rs214087 (c.-166G > C), and rs373592192 (c.-281G > A) variants of NUCB2, suggesting the association of these SNPs with T2D. These findings may support the role of nesfatin-1 or NUCB2 gene in the regulation of blood glucose levels and development of T2D in humans.
In the present study, we genotyped 12 SNPs in the NUCB2 gene in Iranian subjects and assessed the association between NUCB2 and T2DM using a case-control analysis. We demonstrated that some SNPs including rs115148100 (c.-653G > A), rs190662423 (c.-612C > A), rs186174 (c.-427A > C), rs370538176 (c.-426C > G), rs182903196 (c.-308G > A), rs374389403 (c -233T > c), and rs377756452 (c -207G > A) showed no association with T2D.
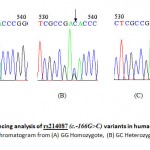 |
Figure 1: Sequencing analysis of rs214087 (c.-166G>C) variants in human NUCB2 gene. A representative chromatogram from (A) GG Homozygote, (B) GC Heterozygote and (C) CC Homozygote.
|
Although the relationship between the promoter region of NUCB2 gene and T2D remains uncertain at present, results of this study further suggest that NUCB2 has an important role in T2D. The nucleotides variants in the promoter region, 5′ UTR and exon 1, the first part of intron 1 of Nucb2 gene, were examined by direct sequencing in patients with T2DM and control subjects, and the data were compared between the two groups. Understanding the relationship between SNP and mutation probability of Nucb2 gene promoter and T2D can be considered as a new therapeutic approach in patients with T2D, such as by reducing the amount of protein used.
Conclusions
Conclusion, we demonstrate, for the first time, the SNPs of the NUCB2 gene promoter region that are associated with T2D. Taken together, studies available so far support an expanding role of NUCB2, thus opening new fields of investigation. However, several questions remain unanswered, in particular, identifying all the SNPs of the promoter region of NUCB2 gene and the strongest SNP associated with T2D. In addition, it is necessary to understand the role of this gene in the development and progression of T2D and find new ways for treatments. Finally, further research on the promoter region of this gene appears to be essential.
Acknowledgements
Our special thanks and gratitude are due to “Neurosciences Research Center (NSRC)” of Tabriz University of Medical Sciences for financial support.
References
- Stengel A, Goebel M, Tache Y. Nesfatin-1: a novel inhibitory regulator of food intake and body weight. Obes Rev. 2011; 12:261-71.
CrossRef - Cao X, Liu Xm Fau – Zhou, Li-Hong, Zhou LH. Recent progress in research on the distribution and function of NUCB2/nesfatin-1 in peripheral tissues. Endocr J. 2013; 60:1021-7.
CrossRef - Stengel A, Tache Y. Minireview: nesfatin-1 an emerging new player in the brain-gut, endocrine, and metabolic axis. Endocrinology. 2011; 152:4033–8.
CrossRef - Aydin S.Multi-functional peptide hormone NUCB2/nesfatin-1. Endocrine.2013; 44:312–5.
CrossRef - Nakata M, Manaka K Fau – Yamamoto S, Yamamoto S Fau – Mori M, Mori M Fau – Yada T, Yada T. Nesfatin-1 enhances glucose-induced insulin secretion by promoting Ca (2+) influx through L-type channels in mouse islet beta-cells. Endocr J. 2011;58: 305-13
CrossRef - Liu F, Yang Q, Gao N, Liu F, Chen S. Decreased Plasma Nesfatin-1 Level Is Related to the Thyroid Dysfunction in Patients with Type 2 Diabetes Mellitus. J Diabetes Res. 2014; 2014: 128014.
CrossRef - Guo Y, Liao Y Fau – Fang G, Fang G Fau – Dong J, Dong J Fau – Li Z, Li Z. Increased nucleobindin-2 (NUCB2) transcriptional activity links the regulation of insulin sensitivity in Type 2 diabetes mellitus. Exp Clin Endocrinol Diabete.2012; 120:91-5
- Stengel A, Goebel-Stengel M, Wang L, Kato I, Mori M, Tache Y. Nesfatin-1(30-59) but not the N- and C-terminal fragments, nesfatin-1(1-29) and nesfatin-1(60-82) injected intracerebroventricularly decreases dark phase food intake by increasing inter-meal intervals in mice. Peptides. 2012; 35:143-8.
CrossRef - Li QC, Wang HY, Chen X, Guan HZ, Jiang ZY. Fasting plasma levels of nesfatin-1 in patients with type 1 and type 2 diabetes mellitus and the nutrient-related fluctuation of nesfatin-1 level in normal humans. Regul Pept. 2010; 159:72-77.
CrossRef - Stengel A, Mori M, Tache Y. The role of nesfatin-1 in the regulation of food intake and body weight: recent developments and future endeavors. Obes Rev. 2013; 14:859-70.
CrossRef - Zhang Z, Li L, Yang M, Liu H, Boden G, Yang G. Increased plasma levels of nesfatin-1 in patients with newly diagnosed type 2 diabetes mellitus. Exp Clin Endocrinol Diabetes. 2012; 120:91-5.
CrossRef - Jr GD, Sherman B, Hosack D, Yang J. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4: P3
CrossRef - Xu J. Wang J. Chen B. SLC30A8 (ZnT8) Variations and type 2 diabetes in the Chinese Han population. Genet Mol Res. 2012; 11:1592-8
CrossRef - Zegers D, Beckers S, Mertens IL, Van Gaal LF, Van Hul W. Association between polymorphisms of the Nesfatin gene, NUCB2, and obesity in men. Mol Genet Metab. 2011; 103:282-6.
CrossRef - Chen YY, Chan R M, Tan K M, Poh LK, Loke KY, Wang JP, et al. The association of a nucleobindin 2 gene (NUCB2) variant with childhood adiposity. Gene. 2013; 516: 48–52.
CrossRef - Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013, 63:11 – 30.
CrossRef - Matteo Riva, Marloes Dekker Nitert, Ulrikke Voss, Ramasri Sathanoori, Andreas Lindqvist, Charlotte Ling, Wierup N: Nesfatin-1 stimulates glucagon and insulin secretion and beta cell NUCB2 is reduced in human type 2 diabetic subjects. Cell Tissue Res (2011) 346:393–405 2011, 346:C393–405.
- Ziru Li, Ling Gao, Hong Tang, Yue Yin, Xinxin Xiang, Yin Li, Jing Zhao, Michael Mulholland, Zhang W: Peripheral effects of nesfatin-1 on glucose homeostasis. PLOS ONE 2013, 8(8).
- Mengliu Yang, Zhihong Zhang, Chong Wang, Ke Li, Shengbing Li, Guenther Boden, Ling Li, Yang aG: Nesfatin-1 action in the brain increases insulin sensitivity through Akt/AMPK/TORC2 pathway in diet-induced insulin resistance. DIABETES 2012, 61:1959-1968.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





