How to Cite | Publication History | PlumX Article Matrix
Mitsuru Akita1, 2 and Manoj Baliram Pohare1*
1The United Graduate School of Agricultural Sciences, Ehime University, 3-5-7 Tarumi, Matsuyama, 790-8566 Japan.
2Graduate School of Agriculture, Ehime University, 3-5-7 Tarumi, Matsuyama, 790-8566 Japan.
Corresponding Author E-mail: b742006b@mails.cc.ehime-u.ac.jp
DOI : http://dx.doi.org/10.13005/bbra/2405
ABSTRACT: Biochemical analyses of protein translocation intermediates formed during early stages of protein import into chloroplasts under restricted energy conditions identified many components involved in protein translocation. Whereas limited information is available at the latter stages of import, as it is difficult to pause the movement of precursor once it was released from the early intermediates. To address this problem, we have attempted to obtain post-early intermediates to plug translocation channel by precursor proteins carrying a tightly-folded structure at their C-termini. To this end, we prepared the recombinant precursor protein whose chloroplastic targeting signal was fused to dihydrofolate reductase from Escherichia coli, known to fold tightly in the presence of its substrate analogue methotrexate. If the precursor was treated with methotrexate prior to the import reaction, the amount of processed precursor was reduced. However, the processed precursor was recovered in the soluble fraction after fractionation, indicating that methotrexate was released from the precursor, which suggested the presence of strong unfolding activity within chloroplasts.
KEYWORDS: protein import into chloroplasts; steric hindrance; recombinant protein; DHFR; methotrexate
Download this article as:| Copy the following to cite this article: Akita M, Pohare M. B. Chloroplastic Protein Import Characteristics of Dihydrofolate Reductase (DHFR) Fused Recombinant Precursor Protein in the Presence of Methotrexate. Biosci Biotech Res Asia 2016;13(4). |
| Copy the following to cite this URL: Akita M, Pohare M. B. Chloroplastic Protein Import Characteristics of Dihydrofolate Reductase (DHFR) Fused Recombinant Precursor Protein in the Presence of Methotrexate. Biosci Biotech Res Asia 2016;13(4). Available from: https://www.biotech-asia.org/?p=17425 |
Introduction
Chloroplasts are the essential cell organelles of the photosynthetic eukaryotes. Apart from their characteristic function, in which it converts light energy into chemical energy through the process of photosynthesis, it is also a home for many other important biosynthetic processes, including synthesis of fatty acids, amino acids and secondary metabolites[1,2]. These functions are governed by ~3000 different proteins present in the chloroplast[3]. But chloroplastic genome encodes for only ~100 chloroplastic proteins[4-6], genes encoding the other chloroplastic proteins were transferred to the nuclear genome during the course of evolution. Under these circumstances, where more than 95 % of the chloroplastic proteins were synthesized on the cytosolic ribosomes[3], chloroplasts have evolved a protein import system[7]. Much knowledge regarding the entire process of protein import into chloroplasts is still unclear and need to be explored soon by considering the importance of chloroplast which feeds most of the living organisms on the earth by producing the food through photosynthesis.
Most of nuclear encoded chloroplastic proteins, which are synthesized as precursors in the cytosol with their amino terminal targeting signal, “transit peptide” are imported into chloroplasts through protein translocation machinery (translocons) embedded in their outer and inner envelope membranes[8-15]. Energy requirements during the process of protein import into chloroplast have divided this process into at least two steps namely ‘docking’ and ‘translocation’[16,17]. In docking, precursor binds irreversibly to the chloroplastic envelope and form the early protein translocation intermediates (PTIs) under limited energy conditions, i.e. low concentrations of GTP and/or ATP, and a low temperature[18-20]. In contrast, translocation requires energy rich conditions (ATP >1 mM), where precursor is imported completely into the stromal space of chloroplasts and was processed by the stromal processing peptidase to produce its mature form[21,22].
Many components consisting translocon identified by biochemical analyses of early PTIs[23-28]. However, limited information is available once after precursors are released from the early PTIs. This is mainly because of the lack of the knowledge regarding the molecular interactions between precursors and the translocon components due to the inability of isolating the PTIs during the translocation step. If the precursors are trapped in the translocon to form PTIs under energy rich conditions (ATP >1 mM), then we can get a way to resolve the molecular interactions during translocation step.
Most common strategy employed to trap the translocating precursor protein in the translocon from different cellular compartments is to fuse a tightly folded domain to the precursor protein which can create the steric hindrance against its import. In mitochondria, precursor protein fused to dihydrofolate reductase (DHFR) has blocked the import once it was stabilized to folded conformation in the presence of methotrexate (MTX)[29]. In case of other membrane systems too, it has been shown that a stable conformation of the precursor protein can block translocation[30,31]. By analyzing the trapped precursor, progress of research on these different cellular compartments was drastically advanced. DHFR—MTX interaction was also applied to chloroplastic protein import research, however, MTX does not block import of precursor protein fused with a tightly folded dihydrofolate reductase (DHFR) from mouse[32,33].
In the current investigation, we made an attempt to reevaluate whether DHFR—MTX interaction did not significantly affect protein import into chloroplasts. Precursors fused to DHFR from mouse (MmDHFR) reported previously was cell-free synthesized incorporating radio-active amino acids without purification, indicating that the amount of precursors were limited and synthesized precursors were contaminated with various cellular factors and small molecules, which might affect blockage of protein import[32,33]. By applying recombinant precursors expressed in E. coli cells, we had developed in vitro chloroplastic protein import assay system[34]. Important features of this assay system are: it can handle the large amount of precursor, import reaction is analyzed without exogenous factors, thus manageable by strict energy conditions. We prepared the new precursor protein fused to DHFR from E. coli (EcDHFR), which was present as a soluble form in E. coli cells after overexpression and retained import competency[35]. DHFR portion of this precursor was tightly folded in the presence of MTX and import of MTX bound precursor was inhibited, though not completely. The results indicate the presence of unfolding activity within chloroplasts, which allow DHFR-fused precursor to be translocated through envelope membranes.
Material and Methods
Construction of plasmids
The expression plasmid harboring the precursor (TP-HA-mSS-H6-BAP, Figure 1A) used as a control for this experiment was modified from the expression plasmid pPsprSSC0HAHAH[34]. The expression plasmid harboring the precursor protein fused to EcDHFR (TP-HA-r1-DHFR-HAT-BAP, Figure 1B) was prepared as described by Pohare and Akita[35]. Furthermore, for the efficient in vivo biotinylation at BAP, the gene for biotin ligase (BirA) from E. coli with ribosome binding site was inserted at the 3’ of the stop codon of the gene for both TP-HA-mSS-H6-BAP as well as TP-HA-r1-DHFR-HAT-BAP on the expression plasmid to form an artificial operon.
Protein expression and purification
Overexpression of both precursor proteins, TP-HA-mSS-H6-BAP and TP-HA-r1-DHFR-HAT-BAP were carried out as described previously except that incubation was for 1 h after induction[36]. After overexpression and cellular fractionation as described previously, precursor proteins were recovered in the insoluble (TP-HA-mSS-H6-BAP) and the soluble (TP-HA-r1-DHFR-HAT-BAP) fractions. The final precipitate containing TP-HA-mSS-H6-BAP was dissolved in solubilization buffer (S-buffer: 8 M urea-25 mM HEPES-KOH, pH 7.5-50 mM KCl-2 mM MgCl2). On the other hand, TP-HA-r1-DHFR-HAT-BAP was purified from the soluble fraction by using Ni-NTA agarose resin (Qiagen) as described previously[35]. Buffer in the eluate was exchanged to import buffer (I-buffer: 50 mM HEPES-KOH, pH 8.0, 330 mM sorbitol) by spin column packed (1 mL bed volume) with Sephadex G-25 (GE Healthcare) as described previously[18].
Protease sensitivity of the precursor
After TP-HA-r1-DHFR-HAT-BAP in I-buffer was incubated with or without 10 µM MTX on ice for 10 min, thermolysin at concentrations indicated was added in the presence of 1 mM CaCl2 and the reaction mixture was incubated at 23 °C for 30 min (Figure 2A). Time-course proteolysis was also carried out with 67 µg mL-1 thermolysin (Figure 2B). Reactions were terminated by adding the sample buffer of Laemmli’s buffer system containing 5 mM EDTA, followed by immediate boiling for 5 min.
Chloroplast isolation and import assay
Chloroplasts were isolated from pea seedlings as described previously[37] and suspended in I-buffer to yield a suspension containing 1 mg of chlorophyll mL-1. When chloroplasts were subjected to the import assay, 20 µL of the chloroplast suspension was preincubated with 2.5 µM nigericin in the dark for 10 min at 25 °C, followed by 4-fold dilution with I-buffer with 1.25 mg mL-1 BSA, 31.25 mM DTT, 6.25 mM MgCl2, and 3.125 mM ATP and incubated in the dark for 5 min at 25 °C to energize chloroplasts. After 16.2 µL of I-buffer and 1.8 µL of 10 µM TP-HA-mSS-H6-BAP solubilized in S-buffer or 18 µL of TP-HA-r1-DHFR-HAT-BAP in I-buffer was mixed with 2 µL of DMSO or 0.1 mM MTX in DMSO and incubated on ice for 10 min, 80 µL of energized chloroplast suspension was added to initiate import reaction and import reaction was carried out by the incubation in the dark for 20 min at 25 °C. The reaction mixture was loaded onto 40 % Percoll in I-buffer and centrifuged (1500 × g) at 4 °C for 5 min to recover intact chloroplasts, which were washed once with I-buffer. Chloroplasts were suspended either in the sample buffer of Laemmli’s buffer system for SDS-PAGE or in HM buffer (25 mM HEPES-KOH, pH 8.0, 2 mM MgCl2) to contain 1 mg of chlorophyll mL-1 to lyse hypotonically.
Fractionation of chloroplasts
Chloroplasts in HM buffer were incubated on ice for 10 min in the dark. After chloroplast suspension was adjusted to contain 0.2 M sucrose by adding HM-2 (HM buffer with 2 M sucrose), chloroplasts were fractionated into the soluble and the membrane fractions by ultracentrifugation as previously described[38].
Electrophoresis and immunoblotting
Proteins in the sample buffer were separated by SDS-PAGE[39] after boiling for 5 min. Once electrophoresis was completed, gels were stained with Coomassie Brilliant Blue (CBB) R-250 or subjected to immunoblotting. Immunoblotting with either anti-HA monoclonal antibody (M180 from MBL) as a primary antibody or streptavidin-AP conjugate (invitrogen) was performed as described previously[34].
Results and Discussion
Precursor development
Knowledge regarding the process of protein import into chloroplasts will definitely expand if one could be able to plug the translocon during the latter stages of import in the presence of high levels of ATP. Steric hindrance created by fusing a tightly folded domain at carboxyl terminal of precursor may plug the translocon. If the precursor with a tightly folded domain is applied for plugging the translocation channel, the precursor shall be in the soluble form. However, all of E. coli overexpressed precursors we prepared so far were recovered in the inclusion bodies and need to be denatured by solubilizing in 8 M urea containing buffer prior to the import reaction[34,36,40-44]. Therefore, firstly we made some efforts in our previous study to develop an import competent EcDHFR fused precursor, TP-HA-r1-DHFR-HAT-BAP (Figure 1B), which was overexpressed in E. coli in a soluble form[35].
 |
Figure 1: Precursor constructs used in this study |
A. TP-HA-mSS-H6-BAP. This precursor was modified from prSSC0HAHAH (Inoue et al., 2008) to possess HA epitope tag which was inserted between fourth and fifth amino acid residue of mature Rubisco small subunit (SS), TEV protease recognition site (TEV), the VSV-G epitope tag, hexahistidine (H6) tag, and biotin acceptor peptide (BAP).
B. TP-HA-r1-DHFR-HAT-BAP. This precursor was modified from TP-HA-mSS-H6-BAP by replacing its mSS-H6 part with random coil linker (r-coil; 4 repeats of GGGGS), EcDHFR and histidine affinity tag (HAT).
Evaluation of MTX-bound precursor
Once TP-HA-r1-DHFR-HAT-BAP was found to be import competent, DHFR portion of this precursor folded tightly in the presence of MTX was evaluated by limited proteolysis with thermolysin. After E. coli overexpressed TP-HA-r1-DHFR-HAT-BAP was purified by Ni-NTA agarose, followed by buffer exchange to I-buffer, protease sensitivity of the precursor was compared depending on the presence of MTX. Thermolysin treated samples were analyzed by immunoblotting decorated with the monoclonal antibody against HA epitope tag (Figure 2). Additional bands appearing below the precursor protein were considered to be the N-terminal truncations of the precursor proteins, as they were appeared after the purification of precursor by using HAT tag which was located towards the C-terminal end of the precursor (Figure 2AB, lanes 1 and 7). Firstly, we tried to determine the least concentration of thermolysin to degrade the precursor without MTX (Figure 2A, lane 4) but not the precursor with MTX (Figure 2A, lane 10) by incubating the precursor solution with different concentrations of thermolysin at 23 °C for 30 min. Secondly, at this concentration of thermolysin (67 µg mL-1), time dependent proteolysis was carried out (Figure 2B). Regardless of MTX treatment, a polypeptide of about 28 kDa was observed when TP-HA-r1-DHFR-HAT-BAP was digested by thermolysin at lower concentrations. This blot was decorated with anti-HA antibody. Furthermore, thermolysin cleaves at the amino terminal side of the following residues, Ile, Leu, Val, Ala, Met, Phe[45], there was no possible recognition site of thermolysin in the random coil linker (4 repeats of GGGGS), this polypeptide contained DHFR. At 67 µg mL-1 of thermolysin, this polypeptide was found to be resistant to proteolysis even in the absence of MTX up to 15 min of reaction (Figure 2B, lanes 2-4). But after 15 min, DHFR was degraded in the absence of MTX (Figure 2B, Lanes 5 and 6), whereas in the presence of MTX, DHFR was still resistant to proteolysis for 30 min (Figure 2B, lanes 8-12). These results clearly indicated that within our newly prepared precursor, DHFR portion was folded during expression and its folding became tight in the presence of MTX.
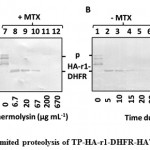 |
Figure 2: Limited proteolysis of TP-HA-r1-DHFR-HAT-BAP |
A. Limited proteolysis of TP-HA-r1-DHFR-HAT-BAP was performed with the indicated concentration of thermolysin (µg mL-1) at 23 °C for 30 min either in the absence or in the presence of MTX.
B. TP-HA-r1-DHFR-HAT-BAP, either in the absence or in the presence of MTX, was treated with 67 µg mL-1 thermolysin at 23 °C for indicated time (min). After proteolysis, samples were separated by SDS-PAGE, followed by immunoblotting decorated with the monoclonal antibody against HA epitope tag. Position of TP-HA-r1-DHFR-HAT-BAP was depicted as “p”. The thermolysin resistant polypeptide which contained a portion of TP-HA-r1-DHFR-HAT-BAP was designated as “HA-r1-DHFR”.
In vitro chloroplastic protein import assay
In order to determine whether engineered precursor protein was targeted to chloroplasts, we employed the in vitro chloroplastic protein import assay with purified TP-HA-r1-DHFR-HAT-BAP (Figure 3A, lanes 7-10) and urea-solubilized TP-HA-mSS-H6-BAP (Figure 3A, lanes 3-6) as a control. In addition, the mock import assay was performed (Figure 3A, lanes 1 and 2). After the import reaction, smaller-sized processed band was produced from both precursors (Figure 3A, lanes 4, 6, 8, and 10). The band appeared around 34 kDa was endogenous chloroplastic biotin binding protein as the same band was observed in the samples of mock assay (Figure 3A, lanes 1 and 2). Import of TP-HA-mSS-H6-BAP was not affected regardless of MTX treatment (Figure 3A, compare lane 4 with 6), indicating that MTX did not affect protein import process. On the other hand, the intensity of the processed band produced from TP-HA-r1-DHFR-HAT-BAP was found to be reduced if this precursor was treated with MTX prior to the import reaction (Figure 3A, compare lane 8 with lane 10). These results suggested that TP-HA-r1-DHFR-HAT-BAP was translocated enough for its cleavage site to be accessible by stromal processing peptidase to cleave its transit peptide to produce the mature form. In addition, less intensity of the imported MTX-treated protein describes the fact that DHFR is tightly folded in the presence of MTX and responsible for its reduced import rate. From these results, however, we were unable to conclude that whether TP-HA-r1-DHFR-HAT-BAP was fully translocated into the stromal space.
Fractionation of chloroplasts
Here, we made an attempt to analyze the nature of the processed proteins by fractionating chloroplasts into the soluble (Figure 3B, lanes 1, 3, 4, 7, 10 and 13) and the membrane (Figure 3B, lanes 2, 4, 5, 8, 11 and 14) fractions after chloroplasts were lysed. The processed band was observed in both fractions in case for TP-HA-mSS-H6-BAP regardless of MTX-treatment (Figure 3B, lanes 4, 5, 7 and 8). Although Rubisco is the soluble protein, substantial amount of endogenous Rubisco was usually recovered in the membrane fraction (data not shown). Since mature part of TP-HA-mSS-H6-BAP was consisted with the mature small subunit of Rubisco, recovery of mature protein produced from TP-HA-mSS-H6-BAP in membrane fraction was highly possible. In the case of TP-HA-r1-DHFR-HAT-BAP, regardless of MTX treatment TP-HA-r1-DHFR-HAT-BAP produced the processed band found in the soluble fraction only (Figure 3B, lanes 10 and 13), indicating that TP-HA-r1-DHFR-HAT-BAP was fully reached to the stromal space. From these results, steric hindrance of tightly folded DHFR by MTX did affect import of TP-HA-r1-DHFR-HAT-BAP, but was not able to keep folded structure. It was previously suggested that strong unfolding activity was present at the surface of chloroplasts[32,33]. Our experimental results also suggested that DHFR became loosely folded or unfolded at the outside of chloroplasts. However, we still have no clue what component is involved in this process.
 |
Figure 3: In vitro chloroplastic protein import assay and fractionation of chloroplasts after protein import |
A. I-buffer (-Precursor), TP-HA-mSS-H6-BAP, and TP-HA-r1-DHFR-HAT-BAP were incubated with (lanes 2, 5, 6, 9 and 10) or without (lanes 1, 3, 4, 7 and 8) MTX before mixed with energized chloroplast suspension. After 20 min incubation at 25 °C, intact chloroplasts were recovered through 40 % Percoll and then washed once with import buffer. Samples were separated by SDS-PAGE and analysed by immunoblotting. The blots were decorated with the streptavidin detecting biotin attached to the BAP tag. Molecular mass (kDa) is shown on the left and the position of TP-HA-mSS-H6-BAP and TP-HA-r1-DHFR-HAT-BAP is depicted as “p”, while processed band is depicted as “m”. 10 % of the precursor applied for the import reaction (lanes 3, 5, 7 and 9) was loaded onto the gel.
B. After import reaction was performed with TP-HA-mSS-H6-BAP, TP-HA-r1-DHFR-HAT-BAP, and buffer only mock sample (-Precursor), chloroplasts were lysed hypotonically and fractionated into the soluble (lanes 1, 3, 4, 7, 10 and 13) and the membrane fractions (lanes 2, 4, 5, 8, 11 and 14). Each fraction was separated with SDS-PAGE (Laemmli, 1970) and analyzed by immunoblotting. The blots were decorated with the streptavidin detecting biotin attached to the BAP tag. Molecular mass (kDa) is shown on the left and the position of TP-HA-mSS-H6-BAP and TP-HA-r1-DHFR-HAT-BAP is depicted as “p”, while processed band is depicted as “m”.
Conclusion
The engineered precursor protein carrying EcDHFR in its mature part in the present study was obtained in the soluble form and found to be import competent. With applying our in vitro chloroplastic protein import assay system free from cytosolic factors, once tightly folded precursor by MTX was imported into chloroplasts at the lesser rate as compared with import of MTX non-treated precursor. This indicates the unfolding activity is present within chloroplasts, regardless of whether this activity is exhibited by an unknown unfoldase present at the surface of chloroplasts or by a mechanical disruption of the tightly folded structure due to the strong pulling force into chloroplasts. If in the latter case, apart from the interest in protein targeting mechanism, to investigate such strong motor, if presents, is valuable for innovating a biological nanomotor.
Acknowledgement
This work was supported by JSPS KAKENHI Grant Numbers 21580415 and 25450132 (to M. A.). This work was carried out at the Division of Genetic Research, the Advanced Research Support Center, Ehime University. M. B. Pohare is a recipient of a scholarship from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Nelson, N., Ben-Shem, A. The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol., 2004; 5: 971–82.
CrossRef - Lopez-Juez, E., Pyke, K.A. Plastids unleashed: their development and their integration in plant development. Int. J. Dev. Biol., 2005; 49: 557–77.
CrossRef - Leister, D. Chloroplast research in the genomic age. Trends Genet., 2003; 19(1): 47–56.
CrossRef - Martin, W., Stoebe, B., Goremykin, V., Hansmann, S., Hasegawa, M., Kowallik, K.V. Gene transfer to the nucleus and the evolution of chloroplasts. Nature, 1998; 393: 162–5.
CrossRef - Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., Leister, D., Stoebe, B., Hasegawa, M., Penny, D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA, 2002; 99(19): 12246–51.
CrossRef - Timmis, J.N., Ayliffe, M.A., Huang, C.Y., Martin, W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet., 2004; 5: 123–35.
CrossRef - Schnell, D.J., Blobel, G., Keegstra, K., Kessler, F., Ko, K., Soll, J. A consensus nomenclature for the protein-import components of the chloroplast envelope. Trends Cell Biol., 1997; 7: 303–4.
CrossRef - Hormann, E., Soll, J., Bolter, B. The chloroplast protein import machinery: A review. Methods Mol. Biol., 2007; 390: 179–93.
CrossRef - Inaba, T., Schnell, D.J. Protein trafficking to chloroplasts: One theme, many variations. Biochem., 2008; 413: 15–28.
CrossRef - Jarvis, P. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol., 2008; 179: 257–85.
CrossRef - Inaba, T. Bilateral communication between plastid and the nucleus: plastid protein import and plastid-to-nucleus retrograde signaling. Biotechnol. Biochem., 2010; 74(3): 471–6.
CrossRef - Shi, L.X., Theg, S.M. The chloroplasts protein import system: from algae to trees. Biophys. Acta, 2013; 1833: 314–31.
- Richardson, L., Paila, Y., Siman, S., Chen, Y., Smith, M., Schnell, D. Targeting and assembly of components of the TOC protein import complex at the chloroplast outer envelope membrane. Plant Sci., 2014; 5: 1–14.
- Ling, Q., Jarvis, P. Functions of plastid protein import and the ubiquitin–proteasome system in plastid development. Biophys. Acta, 2015; 1847: 939–48.
- Bolter, B., Soll, J. Once upon a time–chloroplast protein import research from infancy to future challenges. Plant, 2016; 9: 798–812.
- Chen, X., Schnell, D.J. Protein import into chloroplasts. Trends Cell Biol., 1999; 9: 222–7.
CrossRef - Keegstra, K., Cline, K. Protein import and routing systems of chloroplasts. Plant Cell, 1999; 11: 557–70.
CrossRef - Olsen, L.J., Theg, S.M., Selman, B.R., Keegstra, K. ATP is required for the binding of precursor proteins to chloroplasts. Biol. Chem., 1989; 264: 6724–9.
- Leheny, E.A., Theg, S.M. Apparent inhibition of chloroplast protein import by cold temperatures is due to energetic considerations not membrane fluidity. Plant Cell, 1994; 6: 427–37.
CrossRef - Rensink, W.A., Schnell, D.J., Weisbeek, P.J. The transit sequence of ferredoxin contains different domains for translocation across the outer and inner membrane of the chloroplast envelope. Biol. Chem., 2000; 275: 10265–71.
CrossRef - Chua, N.H., Schmidt, G.W. Post-translational transport into intact chloroplasts of a precursor to the small subunit of ribulose-1,5-bisphosphate carboxylase. Natl. Acad. Sci. USA, 1978; 75: 6110–4.
CrossRef - Lamppa, G.K., Abad, M.S. Processing of a wheat light harvesting chlorophyll a/b protein precursor by a soluble enzyme from higher plant chloroplasts. Cell Biol., 1987; 105: 2641–8.
- Akita, M., Nielsen, E., Keegstra, K. Identification of protein transport complexes in the chloroplastic envelope membranes via chemical crosslinking. Cell Biol., 1997; 136: 983–94.
- Kouranov, A., Schnell, D.J. Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. Cell Biol., 1997; 139: 1677–85.
- Ma, Y., Kouranov, A., LaSala, S.E., Schnell, D.J. Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. Cell Biol., 1996; 134: 315–27.
- Nielsen, E., Akita, M., Davila-Aponte, J., Keegstra, K. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J., 1997; 16: 935–46.
CrossRef - Perry, S.E., Keegstra, K. Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell, 1994; 6: 93–105.
CrossRef - Schnell, D., Kessler, F., Blobel, G. Isolation of the components of the chloroplast import machinery. Science, 1994; 266: 1007–12.
CrossRef - Eilers, M., Schatz, G. Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature, 1986; 322: 228–
CrossRef - Randall, L.L., Hardy, S.J.S. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in coli. Cell, 1986; 46: 921–8.
CrossRef - Muller, G., Zimmermann, R. Import of honeybee prepromellitin into the endoplasmic reticulum: energy requiremerits for membrane insertion. EMBO J., 1988; 7: 639–
CrossRef - America, T., Hageman, J., Guera, A., Rook, F., Archer, K., Keegstra, K., Weisbeek, P. Methotrexate does not block import of a DHFR fusion protein into chloroplasts. Plant Mol. Biol., 1994; 24: 283–94.
CrossRef - Endo, T., Kawakami, M., Goto, A., America, T., Weisbeek, P., Nakai, M. Chloroplast protein Chloroplast envelopes and thylakoids have different abilities to unfold proteins. Eur. J. Biochem., 1994; 225: 403–9.
CrossRef - Inoue, H., Ratnayake, R.M.U., Nonami, H., Akita, M. Development and optimization of an in vitro chloroplastic protein import assay using recombinant proteins. Plant Physiol. Biochem., 2008; 46: 541–
CrossRef - Pohare, M.B., Akita, M. Development of a precursor in a soluble form for protein import into chloroplasts. International Journal of Bio-Technology and Research, 2016; 6(5): 9–
- Inoue, H., Akita, M. Three sets of translocation intermediates are formed during the early stage of protein import into chloroplasts. Biol. Chem., 2008a; 283: 7491–502.
CrossRef - Bruce, B.D., Perry, S., Froehlich, J., Keegstra, K.: In vitro import of proteins into chloroplasts. In: Plant Molecular Biology Manual, Vol. J1, (Gelvin SB, Schilperoort RA, ed). Boston: Kluwer Academic Publishers, 1994: pp 1–15.
CrossRef - Perry, S.E., Li, H.M., Keegstra, K. In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol., 1991; 34: 327–
CrossRef - Laemmli, U.K. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature, 1970; 227: 680–
CrossRef - Akita, M., Inoue, H.: Evaluating the energy-dependent “binding” in the early stage of protein import into chloroplasts. In: Methods in Enzymology, Vol. 466 Biothermodynamics Part B (Johnson ML, Holt JM, Ackers GK, ed). Burlington: Academic Press, 2009; pp 43–
- Inoue, H., Akita, M. The transition of early translocation intermediates in chloroplasts is accompanied by the movement of the targeting signal on the precursor protein. Biochem. Biophys., 2008b; 477: 232–8.
- Ratnayake, R.M.U., Inoue, H., Nonami, H., Akita, M. Alternative processing of Arabidopsis Hsp70 precursors during protein import into chloroplasts. Biotechnol. Biochem., 2008a; 72: 2926–35.
CrossRef - Ratnayake, R.M.U., Kakinuma, Y., Akita, M. Characterization of chloroplast Hsp90 homologue. In: The 2nd International Conference on Bioinformatics and Biomedical Engineering (ICBBE), 2008b: pp 144–
- Sattasuk, K., Inoue, H., Akita, M. In vitro fluorescent analysis of preprotein import into chloroplasts. Biotechnol. Biochem., 2011; 75: 2001–7.
- Keil, B. In: Specificity of proteolysis. Springer-Verlag Berlin-Heidelberg-NewYork, 1992: pp 335.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





