How to Cite | Publication History | PlumX Article Matrix
Ghasem Abbaszadeh-Goudarzi1,2, Kazem Abbaszadeh-Goudarzi1,2, Seyed Davar Siadat2,4٭, Ladan Teimoori-Toolabi1,5, Saeid Bouzari6, Sassan Rezaie3, Mahdi Davari2,4 and M.Reza Khorramizadeh7,1,٭
1Department of Medical Biotechnology, School of Advanced Technologies in Medicine, Tehran University of medical Sciences, Tehran, Iran.
2Department of Mycobacteriology and Pulmonary Research, Pasteur Institute of Iran, Tehran, Iran.
3Department of Medical Mycology and Parasitology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran.
4Microbiology Research Center (MRC), Pasteur Institute of Iran, Tehran, Iran.
5Department of Molecular Medicine, Biotechnology Research Center; Pasteur Institute of Iran, Tehran, Iran.
6Department of Molecular Biology, Pasteur Institute of Iran, Tehran, Iran.
7Biosensor Research Center, Endocrinology and Metabolism Molecular-Cellular Sciences Institute, Tehran University of Medical Sciences, Tehran, Iran.
Corresponding Author E-mail: khoramza@tums.ac.ir
DOI : http://dx.doi.org/10.13005/bbra/2381
ABSTRACT: NontypeableH.influenzae (NTHi) is an important pathogen in children causing otitis media. However,there is no vaccine against NTHi-induced diseases. HMW1,HMW2 and Hia are adhesin proteins of NTHi-strains and have potentiality to provide protection against NTHi-infections. The aim of present study was to construct recombinant HMW1-HMW2-Hia as a fusion protein and evaluate immune responses against recombinant fusion protein as vaccine candidates antigens. Binding domain of hmw1 gene (1080 bp) was amplified by PCR from genomic DNA and cloned into PET-28a vector,this vector was already modified by the insertion of a fragment named hmw2-hia into multiple cloning sites, and then was confirmed by colony-PCR, enzymatic digestion and gene sequencing. To express recombinant fusion protein, PET-28a-hmw1-hmw2-hia vector was transformed into competent BL12(DE3). Expressed protein was purified by affinity chromatography. BALB/c mice were subcutaneously immunized by purified protein in combination with Freund’s adjuvant. Serum antibody responses and functionality of antibodies were determined by ELISA and SBA, respectively. In immunized group of Balb/c mice, the serum IgG responses was significantly increased against recombinant fusion protein in comparison with control, and also the antisera have shown strongly bactericidal activity against NTHi strain. The results have shown that recombinant fusion protein could induce the immune system to produce antibodies which was funtional to kill NTHi-strains. Based on the observation, the antisera have the possibility to provide protection against infections caused by NTHi.
KEYWORDS: NTHi; fusion Protein; Expression Vector; affinity chromatography
Download this article as:| Copy the following to cite this article: Abbaszadeh-Goudarzi G, Abbaszadeh-Goudarzi K, Siadat S. D, Teimoori-Toolabi L, Bouzari S, Rezaie S, Davari M, Khorramizadeh M. R. Cloning, Expression and Purification of Recombinant HMW1 , HMW2 and Hia as a Fusion protein for Vaccine Candidate of NontypeableHaemophilus influenza. Biosci Biotech Res Asia 2016;13(4). |
| Copy the following to cite this URL: Abbaszadeh-Goudarzi G, Abbaszadeh-Goudarzi K, Siadat S. D, Teimoori-Toolabi L, Bouzari S, Rezaie S, Davari M, Khorramizadeh M. R. Cloning, Expression and Purification of Recombinant HMW1 , HMW2 and Hia as a Fusion protein for Vaccine Candidate of NontypeableHaemophilus influenza. Biosci Biotech Res Asia 2016;13(4). Available from: https://www.biotech-asia.org/?p=17082 |
Introduction
Haemophilusinfluenzae is a Gram-negative bacterium that divided to nonencapsulated (nontypeable) and encapsulated (typeable) strains. NontypeableH.influenzae (NTHi) is an important pathogen in children, causing otitis media, sinusitis, conjunctivitis, pneumonia, and typeableH.influenzae strains are responsible for invasive diseases, such as sepsis and meningitis (1). Bacterial adherence to the epithelium of the upper respiratory tract is the first step of colonization and is mediated by both pilus and nonpilus adhesive factors, followed by invasion to the bloodstream or contiguous spread within the respiratory tract(2, 3), and these adhesin proteins have been proposed as vaccine candidates antigens for nontypeableH.influenzae diseases (4).
In nontypeableH.influenzae strains, the HMW1, HMW2 and Hia proteins are major nonpilusadhesins which play an important role in adherence to epithelial cells in a direct way(5, 6). The children who recovered from acute otitis media caused by NTHi strains demonstrated development of the bactericidal antibodies in the sera against highly immunogenic high-molecular-weight (HMW1/HMW2) proteins(7), Subsequently, these studies led to characterization and identification of the HMW1 and HMW2 proteins. Further studies have shown that the HMW1/HMW2 proteins are the main adhesin proteins of nontypeableH.influenza as well as targets of bactericidal and protective antibodies(8, 9). The HMW1/ HMW2 proteins are expressed by nearly 75% of NTHi strains and those NTHi strains (25%) not expressing HMW1/HMW2 proteins have been shown to express a second different class of adhesins known as Hia proteins(9). The Hia proteins have been shown to serve as a target for opsonophagocytic and bactericidalitic antibodies. All of the NTHi strains that lack HMW1/HMW2 proteins contain a hia gene expressing a Hia protein, and vice versa(10).
Binding domain of these proteins are very important for adhesion to the epithelial cells of the respiratory tract for the onset of diseases. The binding domain of HMW1 and HMW2 consist of amino acids residues 555-914 and 553-916 of full-length protein, respectively, and these residues corresponding to amino acids 114-473 and 112 to 475 of mature HMW1/HMW2 proteins in order(11, 12). Hia contain two homologous binding domains, named HiaBD1 and HiaBD2 interacting with the same host cell receptor. The HiaBD1/HiaBD1 is in 585-705 and 51–166 aminoacids of full length protein. Because of the greater affinity of HiaBD1, this binding domain, HiaBD1, was selected as a partner of recombinant fusion protein(13).The purpose of the current study was to produce a recombinant fusion protein which composed of binding domain of HMW1, HMW2, and Hia of nontypeableH.influenzae and to evaluate the antisera productionagainst fusion protein in Balb/c mice. Finallywe aimed, to analyze whether the production of antibodies against recombinant protein is functional or not.
Materials and Methods
Bacterial strains and vectors
NontypeableH.influenzae strain (ATCC 49766) was kindly obtained from the Department of microbiology, Pasteur Institute of Iran. The strain was grown at 37°C on chocolate agar. E. coli Top10 (Invitrogen, USA) and BL-21(DE3) (Novagen, USA) were used as a cloning and expression host, respectively. pTG19-T (Malaysia, Vivantis) and pET-28a (Novagen, USA) were used as a cloning and expression vectors, respectively.
Amplification of binding domain of HMW1555-914
Genomic DNA of the nontypeable H. influenzae strain (ATCC49766) was extracted by high pure PCR template preparation kit (DNA Technology, Russia). Binding domain of hmw1 gene by the length of 1080 bp was amplified by polymerase chain reaction (PCR) from genomic DNA of nontypeableH.influenzae.
To ensure the correct orientation of cloning thethe forward primer (5′-ACTGCCCATGGTTGATGTTCATAAAAAT-3′) contained aNcoI restriction enzyme site, whereas the reverse primer (5′-ACTGCGCTAGCTACATTAAAAGTGAAATT-3′) harbored a NheI site. Amplification of the mentioned gene was achieved by using Pfu DNA polymerase (Thermo scientific, USA), with 1 μl of DNA template (100 ng genomic DNA), 2 μL of 10x reaction buffer, 0.4 μL of dNTPs (10 mM), 0.6 μL of MgCl2 (50 mM), 1 μL of each primer (10 pmol), and 1 U of pfu DNA polymerasein final volume of 20 μL. Hot Start Amplification was performed in 94˚C for 5 min, followed by 30 cycles of denaturation at 94˚C for 30 seconds, annealing at 55˚C for 60 seconds, extension at 72˚C for 120 seconds, and 10 minutes at 72˚C for final extension. The PCR product was recovered from the gel and purified with a high pure PCR product purification kit (Vivantis, Malaysia) based on the manufacturer’s instructions.
Cloning binding domain of hmw1 into PTG19-T vector
Thepolymerized DNA corresponding to the binding domain of hmw1(HMW1 555-914 ) gene was treated with dATP, and then ligated into pTG19-T vector, The ligation was performed by T4 DNA ligase. Recombinant construct as a cloning vector was transformed into competent E. coli TOP10 cells by the CaCl2method. TOP10 cells harboring recombinant vector were grown in LB agar medium supplemented with ampicillin (50 μg/mL). After colony PCR, the recombinant vectors in positive colonies were amplified, and then purified. In order toconfirmation the insertion of hmw1 gene into pTG19-T vector, double digestion by NcoI and NheI restriction enzymes and sequencing were performed.
Subcloning and Expression of Recombinant Fusion Protein
The recombinant cloning vector, rpTG19-HMW1555-914, was digested with NcoI and NheI and the gene corresponding to binding domain of hmw1 ligated into the NcoI- NheI sites of pET28a-hmw2-hiaconstruct, which was already synthesized byas Biomatik, (Canada). Recombinant vectors were transformed into competent E.coli TOP10 cells. After colony PCR, the positive colonies were amplified, and then purified, and finally confirmed by sequencing analysis and double digestion NcoI and NheI. In order to express the recombinant fusion protein HMW1-HMW2-Hia, rpET-28a-hmw1-hmw2-hia, was transformed into competent BL-21(DE3) bacteria. BL21(DE3) cells harboring recombinant vector were grown in LB medium supplemented with kanamycin (50µg/ml) at 37 ◦C with shaking (150 rpm) to reach OD of 0.6in A600. Thereafter, isopropyl β-d-thiogalactopyranoside (IPTG) was added to the final concentration of 1 mM. The BL21(DE3) cells were incubated for 4 hours and then being harvested.
SDS-PAGE and western blotting
The expression of the recombinant fusion protein, HMW1-HMW2-Hia, was analyzed by SDS-PAGE. The bacterial pellets were suspended in a loading buffer sample, heated for 10 minutes at 95°C, and 20 μL of each sample was subjected to 12% SDS-PAGE gel followingby western blotting. The samples were separated by SDS-PAGE method and then proteins on SDS-PAGE gel transferred by wet system to polyvinylidenedifluoride (PVDF) membranes by using a liquid transfer system (Bio-Rad, USA). The membranes were blocked with BSA(2% w/v) in phosphate buffered saline (PBS), and then washed several times with PBS-T. The membranes were incubated with his-tag antibody-HRP conjugate (1:3000 diluted in PBS) for 1 hours at room temperature and developed by diaminobenzidine (DAB) solution (Roche, Germany).
Purification of Recombinant Fusion Protein
A pellet of BL21(DE3) cells harboring recombinant fusion protein, HMW1-HMW2-Hia, were suspended by gentle stirring in lysis buffer (pH 8, containing 300 mMNaCl, 50 mM NaH2Po4, 10 mMImmidazol) followed by freezing, thawing, and sonication on ice in the presence of PMSF (1 mM) as a protease inhibitor. The suspension was subjected to sonication (Six cycles, 30 seconds each, with the intervals of 30 seconds on ice) and then centrifuged for 30 minutes at 10,000 g. After centrifugation to verify the location of the expressed recombinant fusion protein, the precipitate and supernatant were examined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The recombinant fusion protein was purified by affinity chromatography on a nickel-nitrilotriacetic acid (Ni-NTA) gel matrix under denaturing conditions. A pellet of BL-21 cells harboring recombinant fusion protein were suspended by denaturing binding buffer (pH 8, 8 M urea, 100 mM NaH2PO4, 10 mMTris-HCl), and then centrifuged for 30 min at 10,000 g. A column containing 6 ml of Ni-NTA resin was equilibrated with 9 mL of denaturing binding buffer (pH 8, 8 M urea, 100 mM NaH2PO4, and 10 mMTris-Hcl), and then the cleared cell lysate was loaded onto the column. Following adding the samples to the column, it was shook for 60 minutes. The column was washed by washing buffer (pH 6.3, 8 M urea 100mM NaH2PO4, 10mM Tris-Hcl) Finally, The recombinant fusion protein of interest was eluted by elution buffer (pH 5.7, 8 M urea, 100 mM NaH2PO4, 10mM Tris-Hcl). The purified recombinant fusion proteins were pooled and were subjectedto dialysis following a linear gradient of urea from 8 M to 0 M in the refolding buffer (pH 7.2, 20 mM sodium phosphate, and 300 mMNaCl). Finally the protein concentration was determined by Nanodrop spectrophotometry (Thermoscientific,USA). The concentrations of recombinant proteins were determined by Bradford assay with bovine serum albumin (BSA) as the standard protein in NanoDrop spectrophotometry (Bio-RAD, USA)
Mouse immunization
Groups of six-week-old, female BALB/c mice were immunized subcutaneously with 50µg of purified recombinant fusion protein, HMW1-HMW2-Hia, mixed with an equal volume of complete Freund’s adjuvant for the first injection and incomplete Freund’s adjuvant for the second and third injections as a booster14 and 21 days following after the first injection. Control groups of BALB/c mice received Freund’s adjuvant and PBS alone. Antisera were collected 14 and 28 days after injectionto evaluate the antibodyresponses and functionality of those antibodies. Aliquots of antisera were stored at −70 ◦C for assessment.
Determination of serum IgG levels against purified Recombinant Fusion Protein
The IgG responses against purified recombinant fusion proteins,HMW1-HMW2-Hia,weredetermined by ELISA. Serum of female BALB/c mice were collected 14 and 28 days after the first injection, To assess the individual serum samples of immunized and control groups, 96-well flat-bottomed plates were coated overnight at 37 ◦C with 1µg/100 µl of purified recombinant fusion protein,HMW1-HMW2-Hia, in PBS. Following the coating process, the plates were blocked with 1% (w/v) bovine serum albumin (BSA) in PBS for 2 hours at room temperature. After washing, the plates were incubated with antisera (1/256 diluted in PBS) for 2 hours at room temperature. The wells were washed three times with PBS-T thereafter plates were incubated by goat anti-mouse IgG conjugated to Horseradish Peroxidase (Bio-Rad, Hercules, Calif.) in1:3000 dilution in PBS. Plates were incubated for 1 h at 37 ◦C andwashed three times in PBS-T, and then tetramethylbenzidine(TMB) substrate was added to each well. Finally, reaction was stopped after 15 min by adding of H2SO4 2M and the absorbance was measuredat 450 nm. All assays were performed in duplicate.
Serum bactericidal assay
Serum bactericidal assay (SBA) is a functional measurement of the ability of antibodies, in combination with complement, to kill bacteria. Based on the previous protocols, the test was performed(14). Following overnight incubation of nontypeableH.influenzae strain on chocolate agar, single colonies of nontypeableH.influenzae inoculated into LB broth. The NTHi were grown for 5 hours to get to the log phase, it was pelleted by centrifugation and washed and resuspended in PBS containing 9 mMof CaCl2, 4.9 mMof MgCl2 (Dulbecco’s buffer) and 1% (w/v)bovine serum albumin(BSA). In the present study, the SBA was done on pooled serum specimens collected at weeks 0, 2 and 4. All sera were inactivated, by heating for 30 minutes in 56 ◦C all. As a source of complement, 5-week-old serum of baby rabbit was used 10% (v/v). In 96-well, sterile plates SBA assaywas done. The mixture in each well contained 25 µl of complement (20%, v/v), 50 µl of serially diluted sera in buffer, and 25 µl of buffer containing 500 CFU of bacteria, in final volume of 100 µl. After adding components to thewells, microplateswere incubated for 60 minutes at 37 ◦C in a 5% CO2, and then 5 µl aliquot of each well was plated onto a chocolate agar and incubated 16 hours at 37 ◦C. The surviving percentage of bacteria assessed by comparing the respective CFU at 60 minutes compared to those atzero time in the control group. Bactericidal titers defined as the reciprocal of the highest dilution of serum that resulted in ≥50% killing effect in comparison to the control group.Specimens that showed ≤50% bacterial killing effect at the lowest dilution of the tested serum (the lowest dilutionwhich wa tested for serum samples was 1:8) were reported as having a SBA titer of ≤8.All assays were performed in duplicate.
Statistical analysis
All the statistical analysis was done by using SPSS statistical software package (version 17.0). Following normalization of the data, one-way ANOVA and t-tests were used to check for differences between data sets. P values ≤ 0.05 were considered statistically to be significant.
Results
Amplification of hmw1 and the construction of rpET28a-hmw1-hmw2-hia
A 1080bp fragment as a binding domainof HMW1555-914was amplified by PCR using specific primers.Electrophoresis of PCR products showed that the length of HMW1 binding domain was approximately 1080 bp as expected (Figure 1), and thenthis fragment was cloned into pTG19T as a cloning vector.The positive colonies were selected on LB/Amp and existence of HMW1 555-914 insert in recombinant vector,pTG19T-HMW1555-914, was confirmed by colony-PCR, double digestion using NcoI and NheI restriction enzymes (Figure 2,3). The orientation of HMW1555-914in the construct was confirmed by sequencing analysis. Then binding domain of HMW1 555–914,was subcloned into pET28a-hmw2-hia.The positive colonies were selected on LB/Kan and the existenceand the orientation of insert in rPET-28a-hmw1-hmw2-hia, wasconfirmed colony PCR, digestion analysis and sequencing(Figure 4,5).
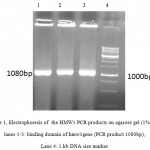 |
Figure 1: Electrophoresis of the HMW1 PCR products on agarose gel (1% w/v) lanes 1-3: binding domain of hmw1gene (PCR product 1080bp), Lane 4: 1 kb DNA size marker. |
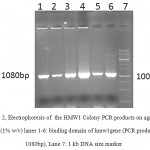 |
Figure 2: Electrophoresis of the HMW1 Colony PCR products on agarose gel (1% w/v) lanes 1-6: binding domain of hmw1gene (PCR product 1080bp), Lane 7: 1 kb DNA size marker. |
 |
Figure 3: Double digestion of the recombinant PTG19-hmw1 vector with NcoI and NheI restriction enzymes., lane 1: : Non digest recombinant PTG19-hmw1 vector , lane 2: Double digestion of the recombinant PTG19_hmw1 (PTG19T: 2880 bp and binding domain of hmw1: 1080 bp), lane 3: Single digestion of the recombinant PTG19-hmw1 (rPTG19-hmw1: 3960 bp)with NcoIenzyme, Lane 4: 1 kb DNA size marker . |
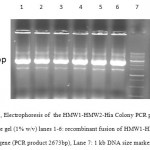 |
Figure 4: Electrophoresis of the HMW1-HMW2-Hia Colony PCR products on agarose gel (1% w/v) lanes 1-6: recombinant fusion of HMW1-HMW2-Hia gene (PCR product 2673bp), Lane 7: 1 kb DNA size marker. |
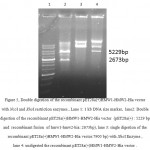 |
Figure 5: Double digestion of the recombinant pET28a(+)HMW1-HMW2-Hia vector with NcoI and XhoI restriction enzymes., Lane 1: 1 kb DNA size marker, lane2: Double digestion of the recombinant pET28a(+)HMW1-HMW2-Hia vector (pET28a(+) : 5229 bp and recombinant fusion of hmw1-hmw2-hia: 2673bp), lane 3: single digestion of the recombinant pET28a(+)HMW1-HMW2-Hia vector:7900 bp)withXhoIEnzyme , lane 4: undigested the recombinant pET28a(+)HMW1-HMW2-Hia vector . |
Expression and purification of recombinant fusion protein HMW1-HMW2-Hia
BL21 (DE3) cells that harbor pET28a-HMW1-HMW2-Hia were cultured at 37 ◦C in the absence and presence of an IPTGwith the concentration of 1 mM. One major band appeared at the 95 kDa position after IPTG induction, as expected at the position of recombinant fusion protein HMW1-HMW2-Hia. Induction of the cells with 1mM IPTG for 4hoursat 37 ◦C,was found to be optimal to achieve high level expression of recombinant fusion protein HMW1-HMW2-Hia. Recombinant fusion protein HMW1-HMW2-Hia was mainly presented in the form of dissoluble inclusion body. Thus, inclusion bodies were carefully purified with Ni-NTA affinity chromatography under denaturing condition The output of recombinant fusion protein HMW1-HMW2-Hiawas approximately 30% of the total bacterial proteins.The highest detectable level of Recombinant fusion protein HMW1-HMW2-Hia was up to 3 mg/L in culturemedume(Figure 6).
 |
Figure 6: SDS-PAGE Analysis of the recombinant fusion protein HMW1-HMW2-Hia protein withCoomassie Staining. Expression and purification of the recombinant fusion proteins in E. coli BL21(DE3). Lane 1,5: protein marker, Lane 2-4: purified recombinant fusion protein marker, Lane 6-8: recombinant fusion protein HMW1-HMW2-Hia induced with IPTG 1mM. |
Western blot analysis
The bands of recombinant fusion protein,HMW1-HMW2-Hia, was detected respectively to be 95 kDa, as expected (Figure 7). The identification of the expressed fusion proteins further confirmed with western blot analysis by using anti-histidine tag monoclonal antibody, where bands corresponding to the molecular mass of the recombinant fusion protein HMW1-HMW2-Hia was respectively detected.
 |
Figure 7: Western Blot Analysis of recombinant fusion purified proteins of HMW1-HMW2-Hia. Lane 1,2: recombinant fusion protein of HMW1-HMW2-Hia with molecular weight of 95kDa , Lane 3: protein marker. |
Serum IgG responses after immunization with recombinant fusion protein
Those BALB/c mice which immunized subcutaneously with recombinantfusion protein,HMW1-HMW2-Hia, mixed with Freund’s adjuvant exhibited high levels of specific antibody responses. Serum IgG responses were significantly increased in immunized groups with recombinant fusion protein, HMW1-HMW2-Hia, in comparison with control groups (P≤0.05). The booster injections were effective to significantly increase the responses of anti-recombinant fusion protein HMW1-HMW2-HiaIgG (Figure 8).
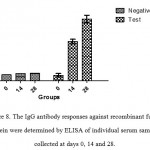 |
Figure 8: The IgG antibody responses against recombinant fusion protein were determined by ELISAof individual serum samples collected at days 0, 14 and 28. |
Serum bactericidal assay
The antiserawere tested for their ability to promote complement-mediated killing of nontypeable H.influenzae (NTHi) strains in vitro. The antisera from immunized animals(Balb/c mice) with recombinant fusion protein,HMW1-HMW2-Hia,were relatively bactericidal against nontypeable H.influenzae (NTHi) strains in comparison with the control group. Antisera raised against recombinant fusion protein,HMW1-HMW2-Hia, demonstrated bactericidal activity with titers of 1/128.
Discussion
Nontypeable H. influenzae (NTHi) strains are responsible for localized respiratory tract diseases such as otitis media, pneumonia and sinusitis.The colonization of nontypeable H. influenza in the upper respiratory is being mediated by adhesin proteins which play an important role in the pathogenesis of diseases(2, 15, 16).Therefore, these adhesin proteins have the potentiality to be utilized as vaccine candidates. Among these proteins, there are a class of proteins named on the basis of their high molecular weight, these adhesin protein are HMW1 and HMW2 and Hia whichare highly immunogenic, surface adhesins and expressed abundant among NTHi strains.The HMW1 and HMW2 proteins are expressed by approximately 75% of nontypeable H.influenzae strains(17, 18).Those strains of Nontypeable H.influenzae strains that do not express HMW1and HMW2 proteins, express immunogenic adhesinsknown as Hia proteins, explaining why fusion proteins should be used for designing a vaccine aiming atprotection against widespread spectrum of diseases caused by NTHi strains(7, 19).
In the present study, we constructed a recombinant fusion protein HMW1-HMW2-Hia from amino acid residues of binding domain of HMW1555–914, HMW2553–916 and Hia585–705, respectively. Evaluation of IgGantibodies have shown the efficacy of vaccine candidate for the induction of immune responses against recombinant fusion protein HMW1-HMW2-Hia. Also serum bactericidal assay(SBA) confirmed that antibodies responses corresponding to immunization with therecombinant fusion protein HMW1-HMW2-Hiawas capable of inducing bactericidal activity against NTHi strains. These data suggest that recombinant fusion protein HMW1, HMW2, and Hia purified as vaccine candidates have a high potential to provide protection against infections caused by NTHi strains.
At this time, there is no vaccine providing protection against NTHi diseases. Our current study has focused on the capability of antisera directed against HMW1-HMW2-Hia fusion proteinwhich also evaluate complement-mediated killing of nontypeableH.influenzae (NTHi) strains in vitro in order to confirm that whether mentioned proteins are good enough to make protection. However,there is a long way to consider these adhesin proteins as good candidatesagainst diseases caused by NTHi. Our present study was adapted frompreviousstudy which demonstrated that a group of common epitopes existing on the HMW1 and HMW2 and Hiaproteins of NTHI could induceactive antibodies (1) and alsothe results of our study coincide with the fact that recombinant adenoviruses expressing recombinant HMW1, HMW2, or Hiaproteins are particularly attractive vaccine candidates providing protection against upper respiratory tract diseases(20). Antibodies against native HMW1, HMW2 proteins and recombinant Hia are suitable of mediating broad-based opsonophagocytic killing of homologous and heterologousNTHi strains(21, 22). A vaccine constructedof HMW1 and HMW2 and Hia proteins might provide protection against diseases caused by many Nontypeable H.influenzae strains. In spite of the fact that binding domain has diversity, it has conserved amino acid motifs which can help us to design a fusion protein as potential vaccine candidates against NTHi.
The results of ELISA and SBA have shown that recombinant fusion protein could induce the immune system to produce antibodies killingnontypeableH.influenzae strain by complement system in vitro. Based on the observation, the antisera against recombinant fusion protein have the possibility to provide a good protection against infections caused by NTHi. Therefore, the results argue for the further investigationdesigning effective vaccines for NTHi-induced diseases based on these proteins or combination of them.
Acknowledgement
This work was financially supported by the Grant No. 18103 ofPh.D Thesis from Tehran University of Medical Sciences, Tehran, Iran. We hereby thank all members of the Department of Mycobacteriology and Pulmonary Research, Pasteur Institute of Iran.
Conflicts of interest
There are no conflicts of interest in this research project.
References
- Winter LE, Barenkamp SJ. Antibodies specific for the Hia adhesion proteins of nontypeable Haemophilus influenzae mediate opsonophagocytic activity. Clinical and Vaccine Immunology. 2009;16(7):1040-6
CrossRef - Meng G, Geme JWS, Waksman G. Repetitive architecture of the Haemophilus influenzae Hia trimeric autotransporter. Journal of molecular biology. 2008;384(4):824-36.
CrossRef - Meng G, Surana NK, St Geme JW, Waksman G. Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. The EMBO journal. 2006;25(11):2297-304.
CrossRef - Foxwell AR, Kyd JM, Cripps AW. Nontypeable Haemophilus influenzae: pathogenesis and prevention. Microbiology and Molecular Biology Reviews. 1998;62(2):294-308.
- Murphy TF. Current and future prospects for a vaccine for nontypeable Haemophilus influenzae. Current infectious disease reports. 2009;11(3):177-82.
CrossRef - Vuong J, Wang X, Theodore JM, Whitmon J, de Leon PG, Mayer LW, et al. Absence of high molecular weight proteins 1 and/or 2 is associated with decreased adherence among non-typeable Haemophilus influenzae clinical isolates. Journal of medical microbiology. 2013;62(11):1649-56.
CrossRef - Winter LE, Barenkamp SJ. Antibodies specific for the high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are opsonophagocytic for both homologous and heterologous strains. Clinical and vaccine immunology. 2006;13(12):1333-42.
CrossRef - Barenkamp SJ. Immunization with high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae modifies experimental otitis media in chinchillas. Infection and immunity. 1996;64(4):1246-51.
- Geme JWS. Molecular and cellular determinants of non‐typeable Haemophilus influenzae adherence and invasion. Cellular microbiology. 2002;4(4):191-200.
CrossRef - Surana NK, Cutter D, Barenkamp SJ, Geme JWS. The Haemophilus influenzae Hia autotransporter contains an unusually short trimeric translocator domain. Journal of Biological Chemistry. 2004;279(15):14679-85.
CrossRef - Dawid S, Grass S, Geme JWS. Mapping of Binding Domains of NontypeableHaemophilus influenzae HMW1 and HMW2 Adhesins. Infection and immunity. 2001;69(1):307-14.
CrossRef - Barenkam S, St Geme J. Identification of surface-exposed B-cell epitopes on high molecular-weight adhesion proteins of nontypeable Haemophilus influenzae. Infection and immunity. 1996;64(8):3032-7.
- Laarmann S, Cutter D, Juehne T, Barenkamp SJ, St Geme JW. The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Molecular microbiology. 2002;46(3):731-43.
CrossRef - Welsch JA, Granoff D. Immunity to Neisseria meningitidis group B in adults despite lack of serum bactericidal antibody. Clinical and Vaccine Immunology. 2007;14(12):1596-602.
CrossRef - Behrouzi A, Bouzari S, Siadat SD, Jafari A, Irani S. Molecular Cloning, Expression and Purification of Truncated hpd Fragment of Haemophilus influenzae in Escherichia coli. Jundishapur journal of microbiology. 2015;8(8).
CrossRef - Geme JWS, Kumar VV, Cutter D, Barenkamp SJ. Prevalence and Distribution of the hmwand hia Genes and the HMW and Hia Adhesins among Genetically Diverse Strains of Nontypeable Haemophilus influenzae. Infection and immunity. 1998;66(1):364-8.
- Geme JWS, Yeo H-J. A prototype two-partner secretion pathway: the Haemophilus influenzae HMW1 and HMW2 adhesin systems. Trends in microbiology. 2009;17(8):355-60.
CrossRef - Giufrè M, Muscillo M, Spigaglia P, Cardines R, Mastrantonio P, Cerquetti M. Conservation and diversity of HMW1 and HMW2 adhesin binding domains among invasive nontypeable Haemophilus influenzae isolates. Infection and immunity. 2006;74(2):1161-70.
CrossRef - Giufrè M, Carattoli A, Cardines R, Mastrantonio P, Cerquetti M. Variation in expression of HMW1 and HMW2 adhesins in invasive nontypeable Haemophilus influenzae isolates. BMC microbiology. 2008;8(1):1.
CrossRef - Winter LE, Barenkamp SJ. Construction and immunogenicity of recombinant adenovirus vaccines expressing the HMW1, HMW2, or Hia adhesion protein of nontypeable Haemophilus influenzae. Clinical and Vaccine Immunology. 2010;17(10):1567-75.
CrossRef - Winter LE, Barenkamp SJ. Antibodies to the HMW1/HMW2 and Hia adhesins of nontypeable Haemophilus influenzae mediate broad-based opsonophagocytic killing of homologous and heterologous strains. Clinical and Vaccine Immunology. 2014;21(5):613-21.
CrossRef - Winter LE, Barenkamp SJ. Naturally Acquired HMW1-and HMW2-Specific Serum Antibodies in Adults and Children Mediate Opsonophagocytic Killing of Nontypeable Haemophilus influenzae. Clinical and Vaccine Immunology. 2016;23(1):37-46.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





