Manuscript accepted on : 05 June 2016
Published online on: --
Plagiarism Check: Yes
Raheleh Norouzi1, Zohreh Hojati2 and Zahra Badr1
1Biology Department, Genetic section, Faculty of Sciences, University of Isfahan, Isfahan, Iran.
2Isfahan University, Department of Biology, Genetics section, Faculty of Sciences, University of Isfahan, Isfahan, Iran.
DOI : http://dx.doi.org/10.13005/bbra/2392
ABSTRACT: Cytokines are a group of signaling glycoproteins that are expressed in respond to stimulus factors and antigens. IFNβ is an important cytokine that play role in immune processes, inflammation and host defense against external factors. In this study, the expression level of IFNβ protein in HEK293T cell line transfected by recombinant construction pBud.IFNβ examine via Dot blot and Elisa protein tests. Sequence of IFNβ gene with using of primers containing Kozak conserved sequence and enzyme restriction site of Kpnl and BglII amplified from pSVMdhfr vector. After treatment by above mentioned enzymes, IFNβ gene cloned in linearized pBud.CE4.1. Structure of recombinant vector were examined via RFLP, Colony PCR and sequencing and finally transformed into competent Ecoli.TOP10 bacteria and next transfected to HEK293T cells. The results of protein tests indicate that recombinant IFNβ gene expressed successfully in HEK293T cell line under eEf1a promoter of selected vector. Proteins produced in prokaryotic systems are lacking glycosylation and thus have different physicochemical properties in comparison with normal protein production in the body. This causes side effects in the patient so the production of IFNβ protein in human HEK293T cells and high expression level of this protein is the benefits of this research.
KEYWORDS: Recombinant interferon beta; HEK293T cell line; pBud.CE4.1; dot blot; ELISA
Download this article as:| Copy the following to cite this article: Norouzi R, Hojati Z, Badr Z. Evaluation of Interferon-Beta Protein Expression in HEK293T Cell-Line Transfected by Recombinant Construction, Pbud.Ifnβ-1a. Biosci Biotech Res Asia 2016;13(4). |
| Copy the following to cite this URL: Norouzi R, Hojati Z, Badr Z. Evaluation of Interferon-Beta Protein Expression in HEK293T Cell-Line Transfected by Recombinant Construction, Pbud.Ifnβ-1a. Biosci Biotech Res Asia 2016;13(4). Available from: https://www.biotech-asia.org/?p=16788 |
Introduction
Interferons are natural cytokines in body. The term “interferon” defines biological activity of soluble materials that interfere with virus replication process. Interferons are expressed in response to a variety of antigens, including Viral RNA, bacterial productions and tumor proteins (1). Generally, interferons are divided into three groups based on amino acid sequences. Group I consists of α, β, ε, δ, ω, κ interferons while the group II only has a member called interferon γ .Group III of interferons has recently been identified and included members λ1, λ2 and λ3 and play an important role in direct response to viral infections(2). Interferons can be divided into three groups based on their cellular origin. Interferon α with the origin of Leukocyte cell (leukocyte interferon), interferon β with the origin of fibroblasts cells (fibroblast interferon) and interferon γ with the origin of lymphocytes cells (immune interferon) are in this division (3 and 4). Interferon α and β in Group I of interferon family have an important role in suppression of viral infections and inflammation (5). Today, two completely different strategies utilize to produce different forms of recombinant interferon beta. IFNβ-Ia form general is the name of drugs produced in CHO cell lines and IFNβ-Ib is the name of drugs that produce with using of E.Coli based expression system. In times past, production of recombinant IFNβ in E. coli cells had some problems such as dimerization and oligomerization of IFNβ resultant from this method which cause isolation of beta-interferon very difficult and time-consuming (4, 6 and 7). In addition, it was found that produced IFNβ by microbial methods always show lower specific activity than the natural form (8). Therefore in the biopharmaceutical industry, mammalian cell culture systems are preferred (9). In this study, expression level of human IFNβ gene will be examined in human embryonic kidney (HEK293T) cell line transfected with the pBud. IFNβ -1a gene structure with using of protein tests such as Dot blot and Elisa.
Materials and Methods
Primer Design and construction of recombinant pBud.IFNβ -1a structure
In order to evaluation of the expression level of IFNβ mRNA, pSVMdhfr vector containing human IFNβ gene was used as a template for PCR. Specific primers were designed with using of the Oligo7 software (Molecular Biology Insight, USA) in a way to allow amplification of the gene coding region of the human IFNβ from pSVMdhfr vector to be provided. In the F and R primers, sequences related to the Kpn I and Bgl II enzymes recognition sites was placed in order to cloning of IFNβ fragment in the pBud.CE4.1 expression vector. Using of GG nucleotides before KpnI restriction site in the forward primers and TC nucleotides after BglII enzyme restriction site in the reverse primer provide possibility of detecting enzyme restriction site. Design of reverse primer is in the way that stop codon TGA is removed with the aim of trace ability of produced protein with using of Anti-His Tag antibody. For this reason and follow-up to preserve of gene reading frame and expression of histidine sequence GA nucleotides placed before KpnI restriction enzyme site. One of the important factors that effect in translation is the presence of proper sequence surrounding translation start codon or the Kozak sequence, so that the amount of synthesized protein of one mRNA is related to the Kozak sequence. Changing of Kozak sequence in order to close it to constant form can help in increasing of translation level. IFNβ gene Kozak sequence in some positions is different from kozak conserve sequence. Kozak constant sequence is in “GCCRCCAUGG” form that R represents a purine base. Kozak Sequence of IFNβ gene is in “GUCAACAUGA” form. So, to close this sequence to constant Kozak sequence, two nucleotides in place -2 and -5 (according to transcription start site) must be changed to C base. Therefore, “GCCACCATGACC” nucleotide sequence related to Kozak conserved form placed in forward primer after KpnI restriction site. Designed primers were prepared by Kowsar Gene-Fanavaran-IRAN Company in lyophilized powder form. PCR reaction was done according to 95 °C Protocol for 6 minutes, followed by 35 cycles of 94 ° C for 30 seconds, 60 ° C for 30 seconds and 72 ° C for 50 seconds and a final cycle 72 ° C for 10 minutes.
FP IFNβ: 5′-GGGGTACCGCCACCATGACCAACAAGTGT-3′
RP IFNβ: 5′-GAAGATCTTCGTTTCGGAGGTAACCTG -3′
The amplified fragment was purified by gel extraction kit (Thermo Scientific, USA). At the same time, pBud.CE4.1 vector was linearized with Kpn I and Bgl II enzymes. At last, amplified fragment cloned in linear vector by T4 DNA ligase enzyme. Ligation mixture was transformed to the competent E. coli.TOP10 bacteria by standard thermal shock method. The colonies containing the recombinant vector were selected on LBA plate with the final 50 μg / ml Zeocin antibiotic concentration. Recombinant colonies verified via Colony PCR method and then plasmid extraction was carried on positive colonies. RFLP analysis with KpnI and BglII enzymes and DNA sequencing with using of Bgh Rev universal primers of Fazabiotech-Iran Company were done.
Cell culture, transfection and protein extraction
After validation of recombinant construction’s structure, HEK293T cells were transfected with using of Lipofectamine® LTX plus™ reagent (Invitrogen, USA) kit. 48 hours after transfection, culture medium was isolated and cells were washed with using of PBS buffer and then were detached from flask with using of 0.05 percent of trypsin. Cells were transferred to a falcon and were centrifuged with 1700 rpm for 5 minutes. After providing cell sediment, different protein extraction process was done with using of TRizol purchased from Life Technologies Company.
Protein detection
Protein was extracted from two phases of cell culture environment (in order to study the possibility of secretory protein IFNβ in the environment) and cell lysis related to transfected cells with recombinant vector, un-transfected control cells and transfected cells with null vector lacking recombinant gene with using of Trizol of Life Technologies with 15596-026 catalog number and according to company instruction. Concentration of obtained proteins were calculated with the help of Bradford standard curve. With the aim of semi-quantitative analysis of protein expression, in each steps, same concentration of proteins were used.
Dot blotting technique
Dot blot is a specific method for identification and semi-quantification analysis of proteins in unknown samples. At this step, a volume equivalent to 24micrograms concentration of extracted proteins from two phases of cell culture environment and cell lysis were loaded onto nitrocellulose paper. 1% SDS protein solvent buffer as negative control and GFP-SCFV protein as positive control were used. After drying at room temperature, the membrane blocked by an appropriate volume of blocking solution (milk powder + PBS) over a night at room temperature with mild agitation on a 95 RPM shaker. Then after three times washing in cold wash buffer (PBS + Tween20) anti-His tagged antibody conjugated with HRP enzyme with proper dilution 1/1000 (4 ml antibody + 4 ml of blocking buffer) used for identification step of recombinant protein. After three washing times in PBS soluble, nitrocellulose papers were treated by this enzyme’s substrate (TMB).
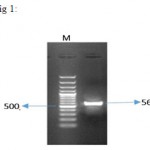 |
Figure 1: result of PCR reaction on pSVMdhfr-IFNβ vector for IFNβ gene amplification. 564 bp fragment of IFNβ was successfully amplified from vector. M represents the size marker with the length of 100 bp (Fermentas).
|
Elisa
ELISA or enzyme-linked immune sorbent assays is a biochemical method that basically is used in order to define the presence of an antibody or an antigen in studied samples. Result of ELISA is a colored reaction which is visible with eyes that has been read with using of multichannel spectrophotometer and data records and analyses, too. In this study, conjugated Monoclonal Anti-poly Histidine antibody with HRP enzyme was used to define concentration of produced IFNβ in HEK293T cell-line. After protein extraction with use of TRizol and determining hole protein concentration via Bradford method, same density equivalent of 25 micro gram in micro liter of all samples and control protein, GFP-SCFV, which has identification sequence of histidine were selected in order to loading in ELISA plate sinks. Selection of same density was for semi-quantitative studies and providing completely same conditions for all samples. SDS buffer which was protein solvent in the last step of protein extraction with the use of TRizol was used as negative control. After observing color change from blue to yellow in samples in result of adding TMB, stop solutions were added and then result read by multichannel spectrophotometer, quickly.
Results
pBud. IFNβ-1a structure construction
PCR of IFNβ gene was performed on PSVMdhfr vector containing the gene with using of designed primers and a product with 564 bp length was amplified (Figure 1). After the cloning process, digestion with two KpnI and BglII enzymes was performed on recombinant vector, and exit of 564 bp IFNβ fragment, confirmed correct entrance of fragment in the pBud.CE4.1 vector (Figure 2).
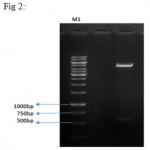 |
Figure 2: double digestion on the recombinant plasmid. Column 1 shows the negative control (uncut vector), column 2 represents linearized vector with a length of 4595 bp and IFNβ gene with a length of 564 bp. M1 is size marker with the length of 1kbp and M2 is the size marker with the length of 100 bp (Fermentas).
|
pBud. IFNβ-1a transfection into HEK293T cell line
Recombinant plasmid was transfected into HEK293T cell-line using Lipofectamine kit (Invitrogen, USA). Moreover, in a separate reaction, the vector without entering segment was transfected into cells. In another flask, un-transfected HEK293T cells were cultured as control cells.
Protein tests
Dot blot reaction was performed to investigate the probability of producing IFNβ protein (Figure 3). In order to final approval of IFNβ protein and performing quantitative and comparative calculations, ELISA reaction was carried out, the results are shown in Figure 4.
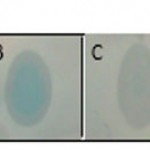 |
Figure 3: representation of produced IFNβ protein in HEK293T cell line using dot blot method. A) Protein extracted from cultured cells transfected with recombinant pBud.IFNβ-1a vector. B) Protein extracted from lysed cells transfected with recombinant pBud.IFNβ-1a vector. C) Positive control protein (GFP-Scfv-His6) D) Negative control (Un-transfected HEK293T cells).
|
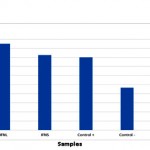 |
Figure 4: Quality comparing of the produced IFNβ protein in HEK293T cells in two phases of cell culture and cell lysis. Positive control is GFP-Scfv protein and negative control are non-transfected HEK293T cells
|
Discussion
Cytokines are cell active factors that are produced in a sustainable manner in response to a wide range of triggers (not only antigens). The significant feature of cytokines is the effect on cellular mechanisms and causing behavioral changes in target cells under certain circumstances (10). Interferons were introduced as the first group of cytokines. Human IFNβ (IFN-β) is a glycoprotein that is widely produced in response to viral infections in fibroblasts cells. That is why it is called fibroblast interferon. IFNβ is used in the field of medicine, in order to treat and improve autoimmune diseases including sclerosis (MS), cell proliferation and angiogenesis inhabitation, and thereby cancer and viral infections treatments. Two analogs of IFNβ are used as drug in the treatment of MS (12). The first type is non-glycosylated form of protein is known as IFNβ-1b which is expressed in E. coli and is currently on the market under the trade name known BetaSeron. Glycosylated form of this protein is IFNβ-1a with the trade names of Avonex and Rebif that both are expressed in ovarian cells of Chinese hamster (CHO). The efficiency of IFNβ-1b produced in bacteria is lower compared to glycosylated form, so that to achieve the efficiency in accordance with the IFNβ-1a, it is necessary to prescribe a higher dose of IFNβ-1b (13). Continuous injection of this drug triggers the production of neutralizing and non-neutralizing antibodies against the IFNβ which inhibits the biological effects of drug and reduce its effectiveness. These factors cause lowering the treatment efficiency and side effects such as flu-like symptoms, liver damage, decreased white blood cells and platelets, depression and headache in patient (14). To overcome these problems, other expression hosts such as yeasts, filamentous fungi, insect cells and plant cells can be used, but these hosts due to different glycosylation pattern from human are less used (15، 16 and 17). HEK293T is a derived cell line from human embryonic kidney cells that grows in cell culture and has been altered to cancer cells via transfecting with adenovirus type 5 DNA. The cells are morphologically similar to epithelial cells and are known as one of the best cell lines for the expression of recombinant proteins, and the production of viral vectors and vaccines in both transient and stable expression since 25 years ago (18, 19). Therefore, in this study HEK293T cells have been used for the expression of recombinant interferon beta.
Conclusion
In this study the expression of recombinant plasmids containing IFNβ gene in HEK293T cell line were reviewed and approved with using of Dot blot and ELISA tests. All tests show successful expression of recombinant IFNβ protein in the selected cell line. This could be due to existence of eEf1a strong promoter in the selected vector and also development of conserved Kozak sequence for IFNβ protein using designed primers for gene cloning. Start speed of translation by ribosome to a large extent depends on the Kozak sequence in the vicinity of the start codon, and the produced protein amounts is dependent on the strength of the sequence (21). In this study, by changing two nucleotide -2 and -5 in Kozak sequence and convert it to conserved form, the protein expression level shows significant increasing compared to basic expression in the cell. Briefly, in a conclusion it can be stated that IFNβ gene expression level has been increased significantly due to cloning in pBud.CE4.1 vector under strong eEf1a promoter. So the selected cell line and vector meet good conditions for producing intended pharmaceutical protein. In addition, changing Kozak sequence of IFNβ gene and convert it to stable and conserved form has significant effect on increasing gene expression level. Considering the transient expression of this protein in fibroblast cells, it is needed to produce more and more interferon protein.
Acknowledgement
This article is produced by using of Ms. Raheleh Norouzi Master’s thesis and research plan (92042856) data. Hereby, the Department of Biology of Isfahan University and the Iran national science foundation are sincerely appreciated for providing facilities and equipment for this study.
References
- Chelbi-Alix, M. K. and Wietzerbin, J. . Interferon, a growing cytokine family: 50 years of interferon research. Elsevier. 2007; (89) 713-718
- Pestka, S., C. D. Krause, et al. “Interferons, interferon like cytokines, and their receptors.”Immunological reviews. 2004; 202(1): 8-32.
CrossRef - Pang, K. . Biological and clinical basis for molecular studies of interferons. Springer. 2005; 116:1
CrossRef - Zago, P., Baralle, M., Ayala, P.M., Skoko, N., Zacchigna, S. . Improving human interferon production in mammalian cell lines by insertion of an intronic sequence within its naturally uninterrupted gene. Wiley Online Library. 2009; (52)191-198
- Friedman, R. M. . Clinical uses of interferons. Wiley Online Library. 2008; (65) 158-162
- Mark, DF, lin, Sl. And Yulu, SD. . Human recombinant cysteine depleted interferon-θ muteins. 1966 US patent 4588585.
- Runkel, L., Meier, W., Pepinsky, R.B., Karpusas, M., Whitty, A., Kimball, K . Structural and functional differences between glycosylated and non-glycosyated forms of human interferon-beta (IFNbeta). Pharmaceutical research. 1998; 15(4): 641-9
CrossRef - Runkel, L. . Structural and functional differences between glycosylated and non-glycosylated forms of human IFNβ (IFN-beta). Springer. 1998; (15)641-649.
- Patrick, H., and sarwat, F. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology. 2009; (19)936-949.
- Morris, A. and I. Zvetkova. “Cytokine research: the interferon paradigm.” Journal of clinical pathology 1997; 50(8): 635-639.
CrossRef - Pestka, S. and Baron, S. . Definition and classification of the interferons. Methods in Enzymology. 1981; (78) 3-14.
CrossRef - Meyer, O. . Interferons and autoimmune disorders. Joint Bone Spine. 2009; 76(5): 464-473.
CrossRef - Bertolotto, A., Deisenhammer, F., Gallo, P., et al. . Immunogenicity of interferon beta: differences among products. Journal of Neurology. 2004; 251(2): II15-II24.
CrossRef - Giovannoni, G., Munschauer, F. and Deisenhammer, F. . Neutralising antibodies to IFNβ during the treatment of multiple sclerosis. Journal of Neurology, Neurosurgery and Psychiatry. 2002; 73(5): 465-469.
CrossRef - Skoko, N., Argamante, B., Grujicić, N. K., et al. . Expression and characterization of human IFNβ in the methylotrophic yeast Pichia pastoris. Biotechnology and Applied Biochemistry. 2003; 38(3): 257-265.
CrossRef - Smith, G. M., Summers , M. D. and Fraser, M. J. . Production of human IFNβ in insect cells infected with a baculovirus expression vector. Molecular and Cellular Biology. 1983; 3(12): 2156-2165.
CrossRef - Squires, C. H. Human Interferon Beta-1b Production in Pseudomonas fluorescens. . BioProcess International. 2010; 8(7): 132.
- Ebtekar, M., K. Azadmanesh, et al. “Cloning Of Human Ifn-1″ (Il-29) From Monocyte Derived Dcs And Its Expression In Hek 293 T.” Arak Medical University Journal (Amuj).
- Thomas, P. and Smart, T. G. HEK293 cell line: a vehicle for the expression of recombinant proteins. Journal of pharmacological and toxicological methods. 200551(3), 187-200.
- Pfaffl, M.W. Quantification strategies in real-time PCR. AZ of quantitative PCR 2004; 1:89-113.
CrossRef - Kozak M, Evans M, Gardner PD, et al. Structural features in eukaryotic mRNAs that mod. Boil Chem. 1991; 2619870-6:19867.

This work is licensed under a Creative Commons Attribution 4.0 International License.





