How to Cite | Publication History | PlumX Article Matrix
Investigation of Calpastatin Gene Polymorphism in Egyptian Sheep and Goat Breeds
Othman E. Othman*, Hassan R. Darwish, Ahmed Abou-Eisha and Adel E. El-Din
Cell Biology Department, National Research Centre, Dokki, Egypt.
Corresponding Author E-mail: othmanmah@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2343
ABSTRACT: Calpastatin is an inhibitor for calpain enzyme system and it has an essential role in meat quality and its tenderness. This trait is of great interest for meat industry and consumers. Molecular genetics techniques help in fishing out of molecular markers which affect the economically production traits in farm animals. Genetic polymorphism of CAST gene and its association with meat quality was reported in different farm animals including cattle, goat and sheep. This work aimed to indentify CAST/MspI genetic polymorphism and its SNPs in Egyptian sheep and goat breeds. The results showed the presence of two genotypes in 140 tested animals, GG and AG with the absence of AA genotypes. The frequencies of GG and AG genotypes in sheep were 65.9% and 34.1%, respectively whereas their frequencies in goat were 56.9% and 43.1%, respectively. The total frequencies for GG and AG genotypes an all 140 tested sheep and goat animals were 62.1% and 37.9%, respectively. The nomenclature of these genotypes was done in this study according to the different nucleotides which were identified after sequencing. The sequence analysis of A and G alleles represented a single nucleotide polymorphism (A→G) at position 286 in the amplified fragment. These nucleotide sequences were submitted to GenBank under the accession numbers, KX722533 and KX722534 (Ovis aries, alleles G and A, respectively) and KX722535 and KX722536 (Capra hircus, alleles G and A, respectively). It is concluded that, CAST genetic polymorphisms in Egyptian sheep and goat breeds were similar to those in other sheep populations around the world where the frequencies of alleles and genotypes with G nucleotide is dominant over the others with A nucleotide. Also, A®G polymorphism in exon 1 of CAST gene is considered a potential molecular marker for marker assisted selection concerning growth rate where the genotypes with G nucleotide is associated with the significant high weight gain in different small ruminant breeds.
KEYWORDS: CAST; PCR-RFLP; DNA sequencing; Sheep; Goat
Download this article as:| Copy the following to cite this article: Othman O. E, Darwish H. R, Abou-Eisha A, El-Din A. E. Investigation of Calpastatin Gene Polymorphism in Egyptian Sheep and Goat Breeds. Biosci Biotech Res Asia 2016;13(4). |
| Copy the following to cite this URL: Othman O. E, Darwish H. R, Abou-Eisha A, El-Din A. E. Investigation of Calpastatin Gene Polymorphism in Egyptian Sheep and Goat Breeds. Biosci Biotech Res Asia 2016;13(4). Available from: https://www.biotech-asia.org/?p=16693 |
Introduction
One of the important characteristics in meat industry and it is of great interest to meat consumers is meat quality. So, the improvement of meat quality and its tenderness is considered one of the essential targets for meat producers (Morgan et al., 1991). Recently, the molecular genetics helps in fishing out of quantitative trait loci which are responsible for economically important production traits. One of QTLs which is reported as an effective marker in meat quality and its tenderness is calpastatin (CAST) (Palmer et al., 1998). Calpain system – which is responsible for skeletal muscle formation and meat tenderness after slaughter – contains μ and m calpain enzymatic molecules and calpastitin is an inhibitor (Goll et al., 1998).
Calpastatin gene is assigned to chromosome 5 in sheep (Huang and Forsberg, 1998) and goat according to the similarity of this chromosome between both species (Evans et al., 1973). Calpastatin can affect muscle protein transformation during animal growth (Forsberg et al., 1989). Many trials were done to resolve the variability in meat quality and its tenderness, some of them were focused on calpastatin and calpain system activity (Bertrand et al., 2001 and Riley et al., 2003). Genetic polymorphism of CAST gene and its association with meat quality was reported in different farm animals including cattle (Schenkel et al., 2006), goat (Khan et al., 2012) and sheep (Chung and Davis, 2012)
In Egypt, there are different major and minor indigenous sheep and goat breeds (Galal et al., 2005 and Abou-Ammou, 2006).These goat and sheep breeds are reared using minimal resources and exposed to compromised and fluctuating environmental production challenges, so they are thought to have acquired unique alleles and allelic combinations that could be important for animal productivity. The contribution of both species to the total red meat produced in Egypt is about 9.1% (MoA, 2004). Sheep and goat meat is favorable to a large scale of population in Egypt especially in rural, desert and Badawi regions, therefore the improvement of meat quality and its tenderness is of great interest in Egypt. For this target, the present work aimed to identify the genetic and single nucleotide polymorphisms of CAST gene in six Egyptian major sheep and goat breeds.
Materials and Methods
Animals and DNA Extraction
The blood samples were collected from one hundred and forty animals belonging to three sheep breeds, Barki (32 animals), Ossimi (28 animals) and Rahmani (22 animals) in addition to three goat breeds, Baladi (16 animals), Barki (20 animals) and Zaraibi (22 animals). Genomic DNA was extracted from the whole blood according to the method described by Miller et al. (1988) with minor modifications. Briefly, blood samples were mixed with cold 2x sucrose-triton and centrifuged at 5000 rpm for 15 min at 4°C. The nuclear pellet was suspended in lysis buffer, sodium dodecyl sulfate and proteinase K and incubated overnight in a shaking water bath at 37°C. Nucleic acids were extracted with saturated NaCl solution. The DNA was picked up and washed in 70% ethanol. The DNA was dissolved in 1X TE buffer. DNA concentration was determined, using Nano Drop1000 Thermo Scientific spectrophotometer, and then diluted to the working concentration of 50 ng/μl, which is suitable for polymerase chain reaction.
Polymerase chain reaction (PCR)
The DNA fragment of the CAST gene was amplified using polymerase chain reaction technique developed by Mullis et al. (1986). A PCR cocktail consists of 1.0 M upper and lower primers (Gharahveysi et al., 2012), 0.2 mM dNTPs and 1.25U of Taq polymerase. The cocktail was aliquot into PCR tubes with 100 ng of sheep or goat DNA. The reaction was cycled with the following conditions, initial denaturation for 5 min at 95°C followed by 35 cycles of denaturation at 95°C (1 min), annealing at 62°C (1 min) and extension at 72°C (2 min) and the final extension for 10 min at 72°C. The amplification was verified by electrophoresis on 2% agarose gel in 1x TBE buffer using GeneRulerTM 100-bp ladder as a molecular weight marker for confirmation of the length of the PCR products. The gel was stained with ethidium bromide and visualized on UV trans-illuminator.
Forward primer: 5¢- TGG GGC CCA ATG ACG CCA TCG ATG -3¢
Reverse primer: 5¢- GGT GGA GCA GCA CTT CTG ATC ACC-3¢
Restriction fragment length polymorphism (RFLP)
Ten μl of PCR product were digested with 1 ul of FastDigest MspI restriction enzymes at 37°C for 5 min. The restriction fragments were subjected to electrophoresis in 2% agarose/ethidium bromide gel (GIBCO, BRL, England) in 1× TBE buffer (0.09 M Tris-boric acid and 0.002 M EDTA). Gels were visualized under UV light and documented in FX Molecular Imager apparatus (BIO-RAD).
Sequence Analysis
The PCR products representing each detected genotype of CAST gene were purified and sequenced by Macrogen Incorporation (Seoul, Korea). Sequence analysis and alignment were carried out using NCBI/BLAST/blastn suite. Results of endouclease restriction were carried out using FastPCR. The nucleotide sequences of different alleles in Egyptian sheep and goat CAST gene were submitted to GenBank (NCBI, BankIt).
Results and Discussion
Advanced molecular genetics techniques have an essential role in animal genetics field during recent decade. These techniques help in the identification of mutations or polymorphisms at DNA level of genes which are associated with economically production traits. Meat quality improvement is considered one of the important targets in meat industry and meat tenderness is an essential characteristic for consumers’ satisfaction (Morgan et al., 1991). One of the problems in this area is the variability of meat tenderness, many trials were done to resolve this problem (Bertrand et al., 2001 and Riley et al., 2003).
Recently, in the way to improvement of meat quality and its tenderness, animal geneticists identified some candidate genes may have effect on meat quality (Sharma et al., 2013). Among these candidate genes, calpastatin is considered one of the promising genes which play an important role in meat tenderness (Li et al., 2010). Calpain system is the endogenous proteolytic system involved in skeletal muscle and meat tenderness. Calpain system includes μ and mcalpain enzymatic molecules in addition to calpastatin as inhibitor, so the increasing of CAST activity is associated with the reduction of meat tenderness (Gharahveysi et al., 2012). This work focused on the identification of genetic and single nucleotide polymorphisms of CAST gene in Egyptian small ruminants toward the improvement of their meat quality through marker-assisted selection.
The amplified fragments from CAST exon 1 at 620-bp (Fig. 1) were digested with MspI endonuclease. Depending on the presence or absence of the restriction site at position 284^285 (C^CGG), the results showed the presence of two genotypes, homozygous GG with two digested fragments at 336- and 284-bp and heterozygous AG with 3 fragments at 620-, 336- and 284-bp (Fig. 2). The nomenclature of these genotypes was done in this study according to the different nucleotides which were identified after sequencing.
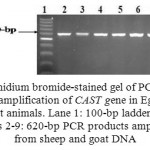 |
Figure 1: Ethidium bromide-stained gel of PCR products representing amplification of CAST gene in Egyptian sheep and goat animals. Lane 1: 100-bp ladder marker. Lanes 2-9: 620-bp PCR products amplified from sheep and goat DNA |
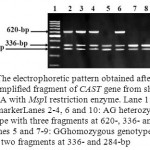 |
Figure 2: The electrophoretic pattern obtained after digestion of PCR amplified fragment of CAST gene from sheep and goat DNA with MspI restriction enzyme. Lane 1: 100-bp ladder marker. Lanes 2-4, 6 and 10: AG heterozygous genotype with three fragments at 620-, 336- and 284-bp. Lanes 5 and 7-9: GG homozygous genotype with two fragments at 336- and 284-bp |
In Egyptian sheep breeds, the frequencies of GG and AG genotypes were 75.0% and 25.0% in Barki, 64.3% and 35.7% in Ossimi and 54.5% and 45.5% in Rahmani with the total frequencies of 65.9% and 34.1% for GG and AG genotypes, respectively. In tested goat animals, the frequencies of GG and AG genotypes were 62.5% and 37.5% in Baladi, 60.0% and 40.0% in Barki and 54.5% and 45.5% in Zaraibi with total frequencies of 56.9% and 43.1% for GG and AG genotypes, respectively (Table 1).The total frequencies for GG and AG genotypes an all 140 tested sheep and goat animals were 62.1% and 37.9%, respectively.
Table 1: Genotype and allele frequencies of calpastatin gene in Egyptian sheep and goat breeds
|
Species |
Breeds |
No. of animals |
Genotype frequencies | Allele frequencies | ||||
| GG | AG | G | A | |||||
| No | Freq | No | Freq | Frequency | Frequency | |||
|
Sheep |
Barki | 32 | 24 | 75.0% | 8 | 25.0% | 87.5% | 12.5% |
| Ossimi | 28 | 18 | 64.3% | 10 | 35.7% | 82.1% | 17.9% | |
| Rahmani | 22 | 12 | 54.5% | 10 | 45.5% | 77.3% | 22.7% | |
| Sub-total | 82 | 54 | 65.9% | 28 | 34.1% | 82.9% | 17.1% | |
|
Goat |
Baladi | 16 | 10 | 62.5% | 6 | 37.5% | 81.25% | 18.75% |
| Barki | 20 | 12 | 60.0% | 8 | 40.0% | 80.0% | 20.0% | |
| Zaraibi | 22 | 12 | 54.5% | 10 | 45.5% | 77.3% | 22.7% | |
| Sub-total | 58 | 33 | 56.9% | 25 | 43.1% | 78.4% | 21.6% | |
| Total | 140 | 87 | 62.1% | 53 | 37.9% | 81.1% | 18.9% | |
These two detected genotypes GG (Fig. 3) and AG (Fig. 4) resulted from the presence of two different alleles A and G. The sequence analysis of these two alleles represented a single nucleotide polymorphism (A→G) at position 286 (Fig. 5) which is responsible for the presence of the restriction site C^CGG at position 284^285 in the allele G. The nucleotide sequences of G and A alleles in Egyptian small ruminants were submitted to GenBank under the accession numbers, KX722533and KX722534 (Ovis aries, alleles G and A, respectively) and KX722535 and KX722536 (Capra hircus, alleles G and A, respectively).
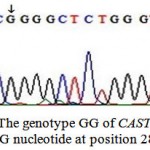 |
Figure 3: The genotype GG of CAST gene with G nucleotide at position 286 |
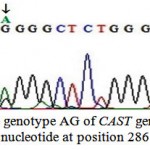 |
Figure 4: The genotype AG of CAST gene with A/G nucleotide at position 286 |
 |
Figure 5: The nucleotide sequence alignment between the twodiferent alleles A and G single nucleotide polymorphism(A→ G) at position 286 in red. |
The identification of CAST polymorphism in Iranian 100 Zel sheep was done using PCR-RFLP technique (Gharahveysi et al., 2012). The digestion of the amplified fragments at 622-bp by MspI restriction enzyme revealed the presence of two alleles M (with G nucleotide) and N with (A nucleotide) and three different genotypes with frequencies, 0.62 (MM), 0.26 (NM) and 0.12 (NN). Genetic structure of CAST gene in Zel sheep was not in Hardy-Weinberg equilibrium (p<0.05) and this gene may be useful in genetic diversity studies in Zel sheep. Also CAST gene polymorphism in Pakistan Thalli, Lohi and Kajli sheep breeds were investigated by Suleman et al. (2012). Frequencies of MM, MN and NN genotypes were found to be 77%, 20% and 3% in Lohi breed and 68%, 26% and 6% in Kajli breed, respectively. In Thalli sheep, only the MM (80%) and MN (20%) genotypes were detected.
Yilmaz et al. (2014) examined the polymorphism of CAST gene in some Turkish sheep breeds. Allele frequencies for M and N alleles of the gene were found to be 0.85 and 0.15 in KIV, 0.80 and 0.20 in KM, 0.99 and 0.01 in GA and 0.34 and 0.66 in SZ sheep, respectively. The effects of CAST genotypes on weight traits in different sheep breeds were investigated by Chung and Davis (2012). RFLP analysis with TaqI restriction enzyme revealed 3 restriction sites and after verification of individual sequences, the difference revealed that substitutions of nucleotides (A to G) at a nucleotide position 208 located in a non-coding region. The results showed that genotypes of CAST were significantly associated with birth weight and average daily gain in tested sheep breeds.
Khan et al. (2012) studied the CAST genetic polymorphism and its association with average daily weight gain in Pakistan sheep and goats. The frequencies of different genotypes were 76% for MM and 24% for MN genotypes in Balkhi sheep breed and 74% for MM, 24% for NM and 2% for NN genotypes in Kajli sheep breed. Whereas, in Beetal goat breed, the only detected genotype was MM with the absence of other two genotypes. The results declared the association between MN genotype with the significant high weight gain in sheep and goats. Another study on CAST polymorphism in New Zealand Boer goat was done by Zhou and Hickford (2008) using PCR-SSCP technique. Seven SSCP patterns resulted from the presence of seven nucleotide substitutions were identified. The result declared non-synonymous amino acid substitution in exon 6 leading to Ser/Arg substitution in domain L of the protein
It is concluded that, CAST genetic polymorphism in Egyptian sheep and goat breeds examined in this work pass along the same line of other studies which were done on small ruminant populations in different geographical locations around the world where the frequencies of alleles and genotypes with G nucleotide is dominant over the others with A nucleotide. Also, A®G polymorphism in exon 1 of CAST gene is considered a potential molecular marker for marker assisted selection concerning growth rate where the genotypes with G nucleotide is associated with the significant high weight gain in different small ruminant breeds.
References
- Abou-Ammou, F.F. Country report on goat production in Egypt. Proceedings Workshop on Recent Advances in Goat Production under Arid Condition, Cairo, 2006, April 10-13, p.p.: 247-254.
- Bertrand, J.K., Green, R.D., Herring, W.O., Moser, D.W. Genetic evaluation for beef carcass traits. J. Anim. Sci., 2001 (Suppl): E190-E200
CrossRef - Chung, H., Davis, M. PCR-RFLP of the ovine calpastatin gene and its association with growth. Asian J. Anim. Vet. Adv., 2012, 7: 641-652.
CrossRef - Evans, H.J., Buckland, R.A., Sumner, A.T. Chromosome homology and heterochromatin in goat, sheep and ox studied by banding techniques. Chromosoma, 1973, 42: 383-402.
CrossRef - Forsberg, N.E., Ilian, M.A., Ali-Bar, A., Cheeke, P.R., Wehr, N.B. Effects of cimaterol on rabbit growth and myofibrillar protein degradation and on calcium dependent proteinase and calpastatin activities in skeletal muscle. J. Anim. Sci., 1989, 67: 3313-3321.
CrossRef - Galal, S., Abdel-Rasoul, F., Anous, M.R., Shaat, I.M. On-station characterization of small ruminant breeds in Egypt. In: Characterization of small ruminant breeds in West Asia and North Africa, 2005, Vol 2, p.p.: 141-193, Luis Inigues (Ed.), ICARDA, Aleppo, Syria.
- Gharahveysi, S., Abbasi, H.A., Irani, M., Abdullahpour, R., Mirhabibi S. Polymorphism investigation of calpastatin gene in Zel sheep population of Iran by PCR-RFLP method. African J. Biotech., 2012, 11: 3211-3214.
- Goll, D.E., Thompson, V.F., Taylor, R.G., Quali, A. The calpain system and skeletal muscle growth. Can. J. Anim. Sci., 1989, 78: 503-512.
CrossRef - Huang, J., Forsberg, N.E. Role of calpain in skeletal-muscle protein degradation. Proc. Natl. Acad. Sci., USA., 1998, 95: 12100-12105.
CrossRef - Khan, S.U.H,, Riaz, M.N., Ghaffar, A., Khan, M.F.U. Calpastatin (CAST) gene polymorphism and its association with average daily weight gain in Balkhi and Kajli sheep and Beetal goat breeds. Pakistan J. Med. Sci., 2012, 44: 377-382.
- Li, J., Zhang, H., Gan, L. Association of CAST gene polymorphisms with carcass and meat quality traits in Chinese commercial cattle herds, Asian_Australian J. Anim. Sci., 2010, 23: 1405-1411
- Miller, S.A., Dykes, D.D., Paletsky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acid Res., 1988, 16(3): 1215-1220.
CrossRef - MoA. Agriculture Economic Sector, Ministry of Agricultur, 2004.
- Morgan, J.B., Savell, J.W., Miller, R.K., Griffin, D.B., Cross, H.R., Shackelford, S.D. National beef tenderness survey. J. Anim. Sci., 1991, 69: 3274-3283
CrossRef - Mullis, K., Facoma, F., Scharf, S., Snikl, R., Horn, G., Erlish, H. Specific amplification of DNA in vitro: the polymerase chain reaction. Cold Sping Harbor Symposium Quantitative Biology, 1986, 51:260.
CrossRef - Palmer, B.R., Roberts, N., Hickford, J.G., Bickerstaffe, R. Rapid communication: PCR-RFLP for MspI and NcoI in the ovine calpastatin gene. J. Anim. Sci., 1998, 76: 1499-1500.
CrossRef - Riley, D.G., Chase, C.C., Hammond, A.C., West, R.L., Johnson, D.D., Olson, T.A., Coleman, S.W. Estimated genetic parameters for palatability traits of steaks from Brahman cattle. J. Anim. Sci., 2003, 81: 54-60.
CrossRef - Schenkel, F.S., Miller, S.P., Jiang, Z., Mandell, I.B., Ye, X., Li, H. Association of a single nucleotide polymorphism in the calpastatin gene with carcass and meat quality traits of beef cattle. J. Anim. Sci., 2006, 84: 291-299.
CrossRef - Sharma, R., Maitra, A., Pandey, A.K., Singh, L.V., Mishra, B.P. Single nucleotide polymorphisms in caprine calpastatin gene. Russian J. Genet., 2013, 49(4): 441-447.
CrossRef - Suleman, M.U., Saeed, M., Khan, R.N., Yousaf, M., Shah, A., Ishaq, R., Ghafoor, A. Calpastatin (CAST) gene polymorphism in Kajli, Lohi and Thalli sheep breeds. African J. Biotech., 2012, 11: 10655-10660.
- Yilmaz, O., Sezenler, T., Ata, N., Yaman, Y. Cemal, I., Karaca, O. Polymorphism of the ovine calpastatin gene in some Turkish sheep breeds. Turkish J. Vet. Anim. Sci., 2014, 38: 354-357.
CrossRef - Zhou, H., Hickford, J.G.H. Allelic polymorphism of the caprine calpastatin (CAST) gene identified by PCR-SSCP. Meat Sci., 2008, 79: 403-405.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





