How to Cite | Publication History | PlumX Article Matrix
Molecular Diagnosis on Glucose 6 Phosphate Dehydrogenase in a Sample of Iraqi Patients
Rehab S. Ramadhan1, Hadeel M. Khalaf1 and Rebah N. Jabber2
1Department of Medical Biotechnology, College of Biotechnology, Al-Nahrain University, Baghdad, Iraq.
2Biotechnology Research Center, Al-Nahrain University, Baghdad, Iraq.
Corresponding Author E-mail: rehabrebah2004@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2354
ABSTRACT: This study was constructed to invertigate Glucose 6 phosphate dehydrogenase deficiency, and the genetic disorder that leading to heamolysis anemia in group of Iraqi patient. A total of 50 blood samples were collected from different hospitals (Yarmook Hospital, Child hospital , AL Alweyaa , and Medical City) these samples varied in severity in of the enzyme deficiency from mild to chronic according to the test of G6PD, and 20 samples served as control (healthy). The period time of samples collection took about three months from March to June 2014. The percentage of mild symptoms was 42% , acute symptoms was 40% and carrier G6PD was 18% as stated on statistics analysis of Chi square. DNA was extracted from samples and subjected to PCR amplification using 3 specific primers designed for purpose of this study, the first primer (Frag I) with product length 115 bp, second primer (FragII) with product length 1500 bp, third primer (Frag III) with product length 2000 bp that amplify the specific site for the carrier and mild patients with G6PD ,while no band was amplified for the chronic patients with G6PD. After comparing cases and with NCBI we found that the percentage insertion was 30% and substituted was 70%.
KEYWORDS: G6PD; type mutation of gene; heamolysis anemia; and molecular diagnosis
Download this article as:| Copy the following to cite this article: Ramadhan R. S, Khalaf H. M, Jabber R. N. Molecular Diagnosis on Glucose 6 Phosphate Dehydrogenase in a Sample of Iraqi Patients. Biosci Biotech Res Asia 2016;13(4). |
| Copy the following to cite this URL: Ramadhan R. S, Khalaf H. M, Jabber R. N. Molecular Diagnosis on Glucose 6 Phosphate Dehydrogenase in a Sample of Iraqi Patients. Biosci Biotech Res Asia 2016;13(4). Available from: https://www.biotech-asia.org/?p=16703 |
Introduction
Glucose-6-phosphate dehydrogenase (G6PD) deficiency is the most common human enzyme defect, being present in more than 400 million people worldwide (Cappellini and Fiorelli, 2008). During glucose 6 phosphate dehydrogenase deficiency, the red cells are unable to regenerate reduced nicotinamide adenine dinucleotide phosphate (NADPH), a reaction that is normally catalyzed by the G6PD enzyme (Ademowo and Falusi, 2002). Since the X chromosome carries the gene for G6PD enzyme, this deficiency mostly affects males. The two major conditions associated with G6PD deficiency are hemolytic anemias and neonatal jaundice, which may result in neurological complications and death (Luzzatto and Gordon-Smith, 2001). Screening and detection of G6PD deficiency helps in reducing such episodes, through appropriate selection of treatment, patient counseling, and abstinence from disease-precipitating drugs such as antimalarials and other agents. G6PD is an enzyme present in the cytoplasm of all cells, acting specifically in the maintenance of the integrity of the erythrocytes, preventing the oxidation of hemoglobin and other cellular proteins so the deficiency of G6PD will cause hemolytic anemia and jaundice induced by ingestion of oxidative drugs and/or broad beans , there are different kinds of hemolysis from mild to severe that are seen to differences in variantsof the disease (Au et al ., 2006). Molecular studies have shown that the G6PD deficiencies are nearly always caused by single amino acid substitutions ( Poggi et al, 1990).
Materials and Methods
Blood Samples were collected from 50 persons suffering from G6PD and, 20 healthy as a control (healthy), their ages ranged between (7 day – 20 years). Samples were subjected to centrifugation at 2000 rpm for 10 min, and was used for DNA extraction. The Samples were obtain from Yarmook Hospital, Child hospital , AL Alweyaa , and Medical City. The collection period from March 2014 to June 2014.
Detection in Glucose 6 phosphate dehydrogenase laboratory
The detection of G6PD was performed using colorimetric kit from Sigma USA according to manufacturer instructions.
DNA Extraction
Total cellular DNA was extracted from blood samples by using the Reliaprep Blood genomic DNA MiniPrep System from Favorgene Taiwan, determination of concatenation and purity of the extracted DNA was done using nanodrop (Techne /UK).
PCR Protocols
Extracted DNA from blood samples and healthy was used in PCR for amplification of FragI, Frag II, Frag III primers table (1) Initial denaturation 94oC for 5 min., 35 cycle contain denaturation 94oC for 1 min, Annealing 60oC,61oC,and 59oC for each gene, extension 72oC for 1 min., and final extension 72oC for 10 min.
Table 1: Primers
| Name | Sequence (5′-3′) | Product length | References. |
| Frag l | F:CTGAAATCTGGCCTCTGTCC
R:GTTCAGCCCCATCTTAGCAG |
115 bp
|
NCBI |
| Frag ll | F:ACCACAAGGTGGCAGCGTTG
R;TGCCTTGCTGGGCCTCGAAGG |
1600 bp
|
NCBI |
| Frag lll | F:CCAGGGACGTGATGCAGAAC
R:GGGCAGGGACATGGACAGTAAGAG |
2000 bp
|
NCBI |
DNA sequencing
Polymerase chain reaction products of G6PD gene using Frag I, II and III for 30 blood samples were sequenced, and aligned with NCBI to examin the presence of SNPs.
Statistical Analysis:
The statistical Package for the social science (SPSS, version 14) was used for statistical analysis of Chi- sequre and deviation from Hardy Weinberg equilibrium distribution of different groups between patients and control, p value < 0.01 was considered statistically significant.
Result and discussion
The distribution of the studied group
Glucose -6-phosphate dehydrogenase (G6PD) deficiency is the common disease. More than 300 variants of G6PD characterized by standard method (Farhud, and Yazdanpanah, 2008).
In this study, fifty sample were collected from patient dignosed with G6PD (males 37, females 13) as shown in table (2).
Table 2: Description of sample study according to gender.
| Gender | Number | Percentage (%) |
| Male | 37 | 74.00 |
| Female | 13 | 26.00 |
| Total | 50 | 100% |
| Chi-square value | — | 11.826 ** |
| ** (P<0.01). | ||
50 samples divided into three groups depending on the severity of the disease as show in figure (1).
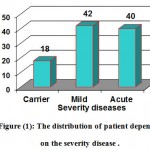 |
Figure 1: The distribution of patient depending on the severity disease |
Molecular detection of G6PD deficiency by PCR technique.
Three primer sets were designed using the NCBI Primer-Design online tool. In order to amplify a specific region in the G6PD gene by using a routine PCR technique.
The G6PD enzyme exits in its active form, as a dimer (or tetramer) each of which consist of 515 amino acid polypeptide subunits (Mason et al , 2007). Each dimer contain tightly bound NADP that plays both structural and functional roles. The enzyme is encoded by G6PD gene, is located on the long arm of the X-chromosome (Xq28) and spans over 18 kilobases consisting of 13 exons and 12 intron (non- coding sequances) (Hesham et al ,2011). The most common mutation occure in exon 7 and frequently detected in Mediterranean, Middle Eastern Hirono and Beutler (1988)
The first primer set used in this PCR technique (Frag I) specific for the intron (6) of G6PD gene from NCBI primer design with product length (115 bp ) which is shown in the figure (1)
 |
Figure 2: Ethidium bromide-stained PCR products after gel electrophoresis. Primer set was used to amplify a sequence of G6PD gene. (2% agarose gel, 10 minutes at 100 voltage and then lowered to 70 Volts, 50 minutes). Negative result was noticed in sample 4,6,10.positive samples were 1,2,3,5,7,8,9 thus indicate successful amplification of the target sequence. The gel also shows a positive control (C), negative control (N), and M represent DNA ladder.
|
 |
Figure 3: Ethidium bromide-stained PCR products after gel electrophoresis. Primer set was used to amplify a sequence of G6PD gene. (1.5% agarose gel, 10 minutes at 100 voltage and then lowered to 70 Volts, 1 h and 30 minutes) Negative result was noticed sample 1,2,3,4,5,7,10. Positive sample were 6,8,9, thus indicate successful amplification of the target sequence. The gel also shows a positive control (C), negative control (N), and M represent DNA ladder
|
The result of this amplification revealed that (70%) were positive and this percentage similar to (Jalloh et al. 2004).
The second primer set used in PCR technique (Frag II) was special to amplify exon 6 and 7 of G6PD gene with product length 1500 bp shown in figure (5).
 |
Figure 4: The automated sequencing of the (G6PD) gene Control (Iraqi healthy)
|
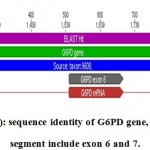 |
Figure 5a: sequence identity of G6PD gene, the amplify segment include exon 6 and 7.
|
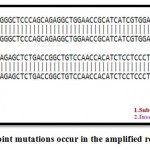 |
Figure 5b: Display point mutations occur in the amplified region in G6PD gene.
|
Four bands were observed in the gel with size nearly 1500 bp. (Ademowo et al., 2002)
The sequncing of the G6PD gene from the all patients in Iraqi populations using primer Frag II and located in exons 6 ,7 and 8.
The amplified sequence in the current study is illustrated in chromosome X, taxon: 9606 in G6PD gene. The segment length was exactly 1049 bp with the upper line (Blast hit) and the sequence amplified started from 1047 – 2095 in the origion NCBI gene include exon 6 with length 159 bp (1548 – 1706) and exon 7 with length 126 bp (1884 – 2009).The green line represent the NCBI control.
Point mutation occurred in the amplified region include transition mutation conversion C/T at position 616, beside to other mutation T insertion occure at position 617. ,( Vulliamy et al.1988) and the same change C / T change at position 563 in exon 6 cause the conversion Ser / Phe at position 188 such conversion might cause loss of enzymefunction and the ability to react normally, other mutation was noticed by (Taki et al. 2001 ) when A/G in exon 6 cause the conversion Asn / Asp was also observed.
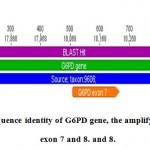 |
Figure 6a: sequence identity of G6PD gene, the amplify segment include exon 7 and 8.
|
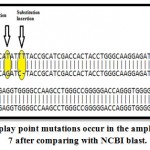 |
Figure 6b: Display point mutations occur in the amplified gene in exon 7 after comparing with NCBI blast.
|
 |
Figure 7: Ethidium bromide-stained PCR products after gel electrophoresis. Primer set was used to amplify a sequence of G6PD gene. (1.5% agarose gel, 10 minutes at 100 voltage and then lowered to 70 Volts, 2h) negative result was noticed in sample 5. Positive samples were 1,2,3,4,6,7,8,9,10,thus indicate successful amplification of the target sequence. The gel also shows a positive control (C), negative control (N), and M represent DNA ladder.
|
In G6PD gene the segment length was amplified exactly 1258 bp start from 18077 – 18235 in the origion NCBI gene include exon 7 with length 159 bp and exon 8 with length 126 bp.
Three point mutation occur in the amplified segment in exon 7, the first was a transversion mutation convert G/T at posision 625, second was a transition mutation C/T at position 628 ,the other mutation was insertion of T at position 629. This variant called the Mediterranean variant , and it has higher affinity for G6PD and reduced thermo stability and associated with acute hemolytic anemia in response to therapeutic drugs and fava beans ( Cappellini , et al, 2008).
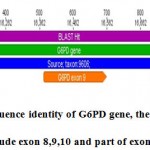 |
Figure 8a: sequence identity of G6PD gene, the amplify segment include exon 8,9,10 and part of exon 11 .
|
 |
Figure 8b: Display point mutations occur in the amplified region in exon 9 after comparing with NCBI blast.
|
The third primer set used in PCR technique (Frag III) exclusively for the exon (9) with product length more than 1000 bp as shown in figure (3-7).
Through four exons length of the amplified segment was exactly 1193 bp start from 15863 – 17055 in the origion NCBI gene include part of exon 8 and complete exon 9 with length 187 bp,exon 10 with length 236 bp start from 16701-16936 at the original fregment and part of exon 11 in G6PD gene .
Point transversion mutation occur when G convert to A at position 650 in exon 9 in G6PD gene. Moradkhani et al. (2012) determine a point mutation in exon (9) of G6PD gene at 1003 when G (guanine ) convert to A (adenine) cause change in amino acid Arg to gln
The overall look for this change might explain the loss of activity or change in stability or in some cases leading to sever enzyme deficiency, enzyme deficiency mostly caused by a point mutation in exon 7 that encoded the most important amino acids for enzyme stability, (Au et al, 2000).
Analysis of G6PD gene by sequencing for Iraqi patients exhibited the existence of many genetic alteration. Two types of mutations were found substitution, and insertion in 70% and 30% respectively. Xu et al. (2005) found the percentage of substitution point mutation in G6PD gene was more than insertion about 70%.
References
- Most of the genetic variants tend to be single-point mutations, and the lack of large or out-of-frame deletions may indicate that the total absence of enzyme activity is fatal . Insertion were usually associated with the most severe clinical manifestations (class I), Mason et al. (2007).
- Ademowo , O.G. ; Falusi , A.G.(2002). Molecular epidemiology and activity of erythrocyte G6PD variants in a homogeneous Nigerian population. East Afr Med J. 79(1):42–44.
CrossRef - Au ,WY; Lam, V; Pang .(2006). Glucose-6-phosphate dehydrogenase deficiency in female octogenarians, nanogenarians, and centenarians. J Gerontol, Ser A, Biol Sci Med Sci .61:1086–9.
CrossRef - Au, S.W. ; Gover, S. ; Lam, V.M. ; Adams, M.J. ; (2000). Human glucose-6-phosphate dehydrogenase: the crystal structure reveals a structural NADP(+) molecule and provides insights into enzyme deficiency. Structure 8: 293–303.
CrossRef - Cappellini, M.D. ; Fiorelli, G. ; (2008). Glucose-6-phosphate dehydrogenase deficiency. Lancet 371, 64–74.
CrossRef - Chan ,TK ; McFadzean , JS. (1974). Haemolytic effect of trimethoprim-sulphamethoxazole in G-6-PD deficiency. Trans R Soc Trop Med Hyg. ; 68(1):61–62.
CrossRef - Farhud, D.D. ; and Yazdanpanah, L. (2008). Glucose-6-Phosphate Dehydrogenase (G6PD) Deficiency. Iranian Journal of Public Health, 37: 1-18.
- Hesham, M. ;Mohammed , S. ; Mohammed , D .(2011). Molecular cloning and characterization of c DNA encoding camelus dromedaries putative G6PD African. J. of Biotech .10(36): 6846-6851.
- Jalloh , A. ; Tantular , IS. ; Pusarawati , S.; Kawilarang, A.P.; Kerong, H.; Lin , K. ; et al. (2004) Rapid epidemiologic assessment of glucose-6-phosphate dehydrogenase deficiency in malaria-endemic areas in Southeast Asia using a novel diagnostic kit. Trop Med Int Health.;9:615–23.
CrossRef - Luzzatto L, Gordon-Smith EC. Inherited haemolytic anaemia. In: Hoffbrand AV, Lewis SM, Tuddenham EGD .(2001) .editors. Postgraduate Haematology. 4th ed. London: Arnold;120–143.
- Martini, G. ; Toniolo, D. ; Vulliamy, T. ; Luzzatto, L. ; Dono, R. ; Viglietto, G. ; Paonessa, G. ; D’Urso, M. ; Persico, M.G., (1986). Structural analysis of the X-linked gene encoding human glucose 6-phosphate dehydrogenase. EMBO J. 5, 1849–1855.
CrossRef - Mason, P.J. ; Bautista, J.M. ; Gilsanz, F., (2007). G6PD deficiency: the genotype-phenotype association. Blood. Rev. 21, 267–283.
CrossRef - Mehta, A.; Mason, P.J. ; Vulliamy, T.J., (2000). Glucose-6-phosphate dehydrogenase deficiency. Bailliere’s Best Pract. Res. Clin. Haematol. 13, 21–38.
CrossRef - Moradkhani, C.; Mekki, M.; Bahuau, V.L.; Te, M.; Holder, S.; Pissard, C.; Préhu, C.; Rose, H.; Wajcman, F. (2012). Practical approach for characterization of glucose 6-phosphate dehydrogenase (G6PD) deficiency in countries with population ethnically heterogeneous: description of seven new G6PD mutants. 208-210.
- Poggi V. ; Town M. ; Foulkes N.S. ; Luzzatto, L. (1990). Identification of a single base change in a new human mutant glucose-6-phosphate dehydrogenase gene by polymerase-chain-reaction amplification of the entire coding region from genomic DNA. Biochem ;271:57–160.
CrossRef - Taki , M.; Hirono, A.; Kawata, M. Den, M.; Kurihara, Y.; Shimizu, H.; Yamada, K.; Fujii ,H.; Miwa, S.(2001) . A new glucose-6-phosphate dehydrogenase variant G6PD Sugao (826C–>T) exhibiting chronic hemolytic anemia with episodes of hemolytic crisis immediately after birth. Int J Hematol , 74:153.
CrossRef - Vulliamy, T.J. ; D’Urso, M. ; Battistuzzi, G. ; Estrada, M. ; Foulkes, N.S. ; Martini, G. ; Calabro, V. ; Poggi, V. ; Giordano, R. Town, M. ; Luzzatto, L. ; Persico, M.G. : (1988).Diverse point mutations in the human glucose-6-phosphate dehydrogenase gene cause enzyme deficiency and mild or severe hemolytic anemia. Proc Natl Acad Sci USA 855171.
CrossRef - Xu, W.; Westwood, B.; Bartsocas, C.S.; Malcorra-Azpiazu, J.J.; Indrák, K.; Beutler, E.(2005). Glucose-6 phosphate dehydrogenase mutations and haplotypes in various ethnic groups, Blood . 257–263.
- Zhao, F. ; Ou, X. L. ; Xu, C. C. ; Cai, G. Q. ; Wu, X. Y. ; Huang, Y. M. ; Zhu, W. F. ; and Jiang, Q. C. (2004). Rapid detection of six common Chinese G6PD mutations by MALDI-TOF MS. Blood Cells Mol. Dis.32: 31.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





