How to Cite | Publication History | PlumX Article Matrix
Tamilvanan1 and V. Murali Bhaskaran2
1Research and Development Centre, Bharathiar University, Coimbatore-641046, Tamil Nadu, India.
2Dhirajlal Gandhi College of Technology, Salem-636290, Tamil Nadu, India.
DOI : http://dx.doi.org/10.13005/bbra/2459
ABSTRACT: In this paper mainly deals with various classification algorithm techniques with feature extraction algorithm used to improve the predicated accuracy of the algorithm. This paper applied with correlation based feature selection as a feature evaluator and Genetics and random searching method. The results of the classification model are sensitive, specificity, precision, time, and accuracy. Finally, it concludes that the proposed CFL-NB algorithm performance is better than other classification algorithms techniques for breast cancer disease.
KEYWORDS: Correlation-based Feature Selection; Data Mining; Genetic Algorithm; Random Search and Naive Bayes Algorithm
Download this article as:| Copy the following to cite this article: Tamilvanan T, Bhaskaran V. M. A New Feature Selection Techniques Using Genetics Search and Random Search Approaches for Breast Cancer. Biosci Biotechnol Res Asia 2017;14(1) |
| Copy the following to cite this URL: Tamilvanan T, Bhaskaran V. M. A New Feature Selection Techniques Using Genetics Search and Random Search Approaches for Breast Cancer. Biosci Biotechnol Res Asia 2017;14(1) Available from: https://www.biotech-asia.org/?p=21840 |
Introduction
Data mining techniques and application are utilized in a wide range of fields, including banking, gregarious science, inculcation, business industries, bioinformatics, weather, forecasting healthcare and sizably voluminous data. Nowadays health care industry generates a large amount of data about patients, disease diagnosis, etc. Some different types of approaches to building accurate classifications have been proposed (e.g., NB, MLP, SMO, RF). In classification, we give a Breast Cancer data set of example record or the input data, called the test data set, with each record consisting of various attributes
An attribute can be either a numerical attribute or categorical attribute. If values of an attributes belong to an authoritatively mandated domain, the attribute is called numerical attribute ( e.g. Age, Menopause, Tumor-size, Inv-nodes, Deg-Malig). A categorical attribute (e.g.Node-Capes, Breast, Breast-Quad, Irradiat, Class). Classification is the way toward part a dataset into totally unrelated gatherings, called a class, based on suitable attributes.
This paper is organized accordingly: the relates works and depiction of the specialized parts of the utilized information mining techniques in section 1. The elaborates with classification algorithms like navie bayes, multi-layer perception, Sequential minimal optimization and random forest in section 2.The introduction of the dataset for Breast Cancer in section 3. The Experiment Results and Discussion in section 4. And finally, conclude the paper and future works.
Correlation Based Feature Selection
Correlation based feature selection9,10,18 is one of the notable methods to rank the pertinence of elements by measuring amongst elements and classes and amongst elements and different components. Given number of components k and classes c, CFS portrayed centrality of parts subset by using Pearson’s relationship condition
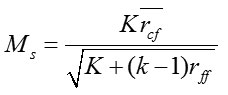
Where Ms is the significance of highlight subset
![]()
is the normal direct correlation coefficient between these elements and classes and
![]()
is the normal straight correlation coefficient between various elements.
Typically, CFS includes (forward choice) or erases (in reverse determination) one element at once. in this work we used Genetic search and Random search algorithms for the great results.19,20
Genetic Search Algorithm
Look strategies explore the ascribe space to find a not too bad subset and the quality is measured by the property subset evaluator through CFS subset evaluator and genetic search is being used as a request technique. The parameters of the genetic algorithm are various generations, people appraise and the probabilities of change and hybrid. A person from the basic masses makes by deciding a rundown of value records as a hunt point. For delivering progress reports, each such an assortment of generations can be Utilized.20,21 The basic genetic search procedure is demonstrated as follows:
Step 1: Start by randomly generating an initial people
Step 2: Calculate e(x) for each fellow xεR.
Step 3: Define a probability distribution p over the Fellows of R where p(x) ∞ e(x).
Step 4: Choose two population members x and y to produce new population members x’ and y’.
Step 5: Apply mutation to x’ and y’.
Step 6: Insert x’ and y’ into P’
Step 7: If |R’| < |R|, go to step 4.
Step 8: Let R ßR’.
Step 9: If there are more generations to process, go to step 2.
Step 10: Return x ε P for which e(x) is highest.
Random Search Algorithm
Step 1: Set algorithm parameters Θ0, initial points X0 ⊂ S And iteration index k = 0.
Step 2: Generate a collection of candidate points Vk+1 ⊂ S according to a specific generator and associated Sampling distribution.
Step 3: Update Xk+1 based on the candidate points Vk+1, Previous iterates, and algorithmic parameters. Also, update algorithm parameters Θk+1.
Step 4: If a stopping criterion is met, stop. Otherwise Increment k and return to Step 1.
Naive Bayes Classification
Naive Bayes27-30 executes probabilistic naive Bayes classifier. naive means restrictive autonomy among traits of components. The “naive” supposition incredibly diminishes calculation unpredictability to a basic increase of probabilities. The Naive Bayes handles numeric properties utilizing directed discretization and utilizations piece thickness estimators that will enhance the execution It requires just little arrangement of preparing information to create exact parameter estimations since it requires just the computation of the frequencies of characteristics and property result combines in the preparation information set30,31
Dataset
The dataset utilized as a part of this model gathered from UCI machine learning repository32 ought to be more exact what’s more, exact remembering the true objective to upgrade the judicious precision of data mining errands. The data set may have missing (or) irrelevant attributes and these are to be dealt with successfully by the data mining process.
Attribute Identification
The breast cancer dataset which comprises of 286 instances and 10 attributes with the class expressing the life visualization yes (or) no. appear in Table 1.
Table 1: Dataset for Breast cancer
| Attributes Name | Description |
| Age | Age (years) |
| Inv-Nodes | 0-2, 3-5, 6-8, 9-11, 12-14, 15-17, 18-20, 21-23, 24-26, 27-29, 30-32, 33-35, 36-39 |
| Node-Caps | yes, no |
| Menopause | lt40, ge40, premeno |
| Tumor-Size | 0-4, 5-9, 10-14, 15-19, 20-24, 25-29, 30-34, 35-39, 40-44,45-49, 50-54, 55-59 |
| Deg-Malig | 1, 2, 3. |
| Breast | left, right |
| Breast-Quad | left-up, left-low, right-up, right-low, central. |
| Irradiat | yes, no. |
| Class | no-recurrence-events, recurrence-events |
Methodology of Proposed Systems
The exploration procedure has two stages. First one is genetics algorithm based CFS connected to the breast cancer dataset which was reduced to 5 from 10 and the Naive Bayes algorithm connected for classification. The second phase of this work utilized Random search based CFS connected to the same data set which was reduced to 6 from 10 and after that connected Naïve Bayes classification algorithm for better expectation.
Experimental Results and Discussion
The exploratory outcomes outline the distinctive measures that are used to evaluate the model for characterization and desire. In this work the specificity, precision, sensitivity and accuracy are expounded.
Sensitivity, Accuracy, Specificity and Precision
Accuracy = (TP + TN)/(TP + TN + FP + FN) (1)
Sensitivity = TP/(TP + FN) (2)
Specificity = TN/(TN + FP) (3)
Precision = TP/(TP + FP) (4)
Table 2: Before Feature Selection
| Classifier | Precision | Sensitivity | Specificity | Time | Accuracy |
| NB | 83% | 77% | 52% | 0.00 | 71% |
| SMO | 85% | 75% | 48% | 0.09 | 69% |
| MLP | 74% | 75% | 40% | 2.18 | 64% |
| RF | 86% | 74% | 48% | 0.1 | 69% |
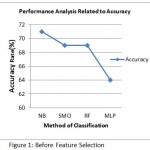 |
Figure 1: Before Feature Selection
|
Table 3: After Feature Selection using Genetics Search
| Classifier | Precision | Sensitivity | Specificity | Time | Accuracy |
| NB | 83% | 78% | 54% | 0.00 | 72% |
| SMO | 81% | 73% | 41% | 0.01 | 66% |
| MLP | 84% | 77% | 53% | 0.99 | 71% |
| RF | 88% | 74% | 50% | 0.03 | 70% |
Table 4: After Feature Selection using Random search
| Classifier | Precision | Sensitivity | Specificity | Time | Accuracy |
| NB | 39% | 51% | 82% | 0.00 | 77% |
| SMO | 30% | 53% | 80% | 0.06 | 76% |
| MLP | 32% | 41% | 80% | 1.11 | 73% |
| RF | 33% | 52% | 81% | 0.13 | 76% |
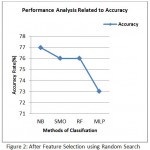 |
Figure 2: After Feature Selection using Random Search
|
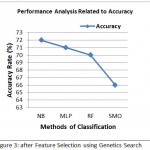 |
Figure 3: after Feature Selection using Genetics Search
|
10-Fold Cross-Validation
The classification algorithm is arranged and attempted in 10 times. The cross approval isolates the information into 10 subgroups and each subgroup is attempted through request oversee worked from whatever is left of the 9 bundles. Ten unmistakable test results are gotten for each train–test setup and the typical result gives the test exactness of the calculation.
Confusion Matrix
The confusion matrix7 plots what quantities of cases have been apportioned to each class and the parts of the lattice speak to the amount of experiments whose genuine class is the line and whose foreseen class is the portion. Tables 5, 6 and 7 depict the perplexity matrix that is processed for Naive Bayes and the innate interest based CFS-NB and sporadic chase based CFS-NB computations.
Table 5: Before Feature Selection based on Naive Bayes
| a | b | Classified as |
| 168 | 33 | a = no-recurrence-events |
| 48 | 37 | b = recurrence-events |
Table 6: After Feature Selection for Genetic search Based on Naive Bayes
| a | b | Classified as |
| 27 | 41 | a = Yes |
| 27 | 191 | b = No |
Table 7: After Feature Selection for Random search Based on Naive Bayes
| a | b | Classified as |
| 27 | 41 | a = Yes |
| 25 | 193 | b = No |
Graph Results
Figure 3 demonstrate the precision of different grouping calculations that was accomplished through genetic search based CFS. Figure 2 demonstrate the precision of different order calculations that was accomplished through random search based CFS.
Conclusion
In this work, an enhanced technique was made for breast cancer analysis. The results exhibit that Random search based CFS-NB achieved comparable characterization correctnesses for a decreased component subset that contained six elements. The genetic search based CFS-NB was conveyed better arrangement precision for a decreased subset of five elements. The relative review was coordinated on the breast cancer information in light of random and genetic search in view of CFS with other characterization calculations like Multilayer perceptron and Sequential Minimal Optimization and Random Forest. The trial result simply portrays that the genetic search based CFS and naive bayes execution was better contrasted and other grouping calculations as far as time and accuracy.
References
- Yua Q., Tan K. C., Toeh E. J., Goh K. C. A hybrid evolutionary algorithm for attribute selection in data mining. Expert Systems with Applications. 2009;36:8616–30.
CrossRef - Kim J. S., Yoon H., Park C. S., Baek J. G. Algorithm learning based neural network integrating feature selection and classification. Expert Systems with Applications. 2013;40:231–41.
CrossRef - Sahin F., Chandrasekar G. A survey on feature selection methods. Computers and Electrical Engineering. 2014;40:16–24.
CrossRef - Gupta M., Chandra B. An efficient statistical feature selection Approach for classification of gene expression data.Journal of Bio medical Informatics. 2011;44:529–35.
CrossRef - Yu L., Liu H. Toward Integrating Feature Selection Algorithms for Classification and Clustering. IEEE Transactions on Knowledge and Data Engineering. 2005;17(4).
- Krishnakumari P., Purusothaman G. A Survey of Data Mining Techniques on Risk Prediction: Heart Disease. Indian Journal of Science and Technology. 2015;8(12).
- Sumathi C. P., Suganya P. A Novel Metaheuristic Data Mining Algorithm for the Detection and Classification of Parkinson Disease. Indian Journal of Science and Technology. 2015;8(14).
- Nasira G. M., Kalaiselvi C. Prediction of Heart Diseases and Cancer in Diabetic Patients Using Data Mining Techniques.Indian Journal of Science and Technology. 2015;8(14).
- Smith L. A.,Hall M. A. Feature Selection for Machine Learning: Comparing a Correlation based filter approach to the wrapper. FCAIRS Conference. 1999.
- Hall M. Correlation-based feature selection for discrete and numeric class machine learning. Proceedings of the Seventeenth International Conference on Machine Learning. 2000;359–66.
- Deng Y., Lu X., Peng X., Feng B., Liu Ping, Liao B. A Novel Feature Selection Method Based on Correlation-Based Feature Selection in Cancer Recognition. Journal of computational and Theoretical nano science. 2014;11(2):427– 33.
CrossRef - Kennedy J., Eberhart R. C. A new optimizer using particle swarm theory. In Proceedings of the sixth international symposium on micro machine and human science. 1995;1:39–43.
- Eberhart R. C., Shi Y. Empirical study of particle swarm optimization. IEEE Proceedings of the 1999 Congress on Evolutionary Computation. 1999;3.
- Eberhart R. C., Shi Y. Particle swarm optimization: developments, applications and resources. IEEE Proceedings of the 2001 Congress on in Evolutionary Computation. 2001;1:81–6.
CrossRef - Sudha S., Vijayarani S. An Efficient Clustering Algorithm for Predicting Diseases from Hemogram Blood Test Samples. Indian Journal of Science and Technology. 2015;8(17).
- García-Saez G., Caballero-Ruiz E., Balsells M., Pons B., Morillo M., Gomez E. J., Hernando M. E. Automatic Blood Glucose Classification for Gestational Diabetes with Feature Selection: Decision Trees vs. Neural Networks. XIII Mediterranean Conference on Medical and Biological Engineering and Computing, 2013. IFMBE Proceedings. 2014; 41:1370–3.
- Gao Y., Xu J., Sun L. X. T. An ensemble feature selection technique for cancer recognition. Bio-Medical Materials and Engineering. 2014;24:1001–8.
- Murugan A., Sridevi T. A Novel Feature Selection Method for Effective Breast Cancer Diagnosis and Prognosis. International Journal of Computer Applications. 2014;88(11).
- Yi Y., Wang J., Zhou S., Kong J. An Improved Feature Selection Based on Effective Range for classification. The Scientific World Journal. 2014;1–8.
- Yang A., Zang Y., Xiong C., Wang T., Zhang Z. Feature Selection for Data Envelopment Analysis. Knowledge Based Systems. 2014;64:70–80.
CrossRef - Li Y., Ding S., Shi Z., Yan S. A Protein Structural Classes Prediction Method based on Predicted Secondary Structure and PST-BLAST Profile. Biochimie. 2014;97:60-5.
CrossRef - Kennedy J., Riccardo P., Blackwell T. Particle swarm optimization.Swarm intelligence. Kennedy J. Particle swarm optimization. 2007;1(1):33–57. Encyclopedia of Machine Learning. Springer US. 2010;760–6.
- Mukhopadhyay A., Mandal M. A novel PSO-based graphtheoretic approach for identifying most relevant and non-redundant gene markers from gene expression data. International Journal of Parallel, Emergent and Distributed Systems. 2014;1–18.
- Wang B. , Ji Z. Identifying potential clinical syndromes of Hepatocellular carcinoma Using PSO-based hierarchical feature selection algorithm. Bio Med research international. 2014;1–12.
- Ananthanarayanan N. R.,Yasodha P. Analyzing Big Data to Build Knowledge Based System for Early Detection of Ovarian Cancer. Indian Journal of Science and Technology.2015;8(14).
- Dumitru D. Prediction of recurrent events in breast cancer using the Naive Bayesian classification. Annals of University of Craiova Math Comp Sci Ser. 2009;36(2):92–6.
- Chandrasekaran R. N., Nagarajan S. Design and Implementation of Expert Clinical System for Diagnosing Diabetes using Data Mining Techniques. Indian Journal of Science and Technology. 2015;8(8):771–6.
CrossRef - Velmurugan T., Anuradha C. A Comparative Analysis on the Evaluation of Classification Algorithms in the Prediction of Students Performance. Indian Journal of Science and Technology. 2015;8(15).
- Lee K. H., Leung K. S., Wang J. F., Ng E. Y., Chan H. L., Tsui S. K.,Mok T. S., Tse p. C., Sung J. J. Data Mining on DNA Sequences of Hepatitis B Virus. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2011;8(2):428–40.
CrossRef - Thangaraju P., Karthikeyan T. Analysis of Classification Algorithms Applied to Hepatitis Patients. International Journal of Computer Applications. 2013;62(5):25–30.
- Available from:http://archieve.ics.uci.edu/ml/datasets/ Breast Cancer/ Accessed on: 2015;27:5.
- Thangaraju P., Karthikeyan T. PCA-NB Algorithm to Enhance the Predictive Accuracy. International Journal of Engineering and Technology. 2014;6(1):381–7.
- Thangaraju P., Karthikeyan T. A Combined Approach of CFS and Naive bayes Algorithm to Enhance the Prediction Accuracy. IEEE International Conference on Control Instrumentation Communication and Computational Technologies (ICCICCT 2014). 2014.
- Thangaraju P., Karthikeyan T. Best First and Greedy Search based CFS-Naive Bayes Classification Algorithms for Hepatitis Diagnosis. Biosciences and Biotechnology Research Asia. 2015;12(1):983–90.
CrossRef - Frank E., Written I. H. Data mining Practical Machine Learning Tools and Techniques. Morgan Kaufmann Publishers. 2011.

This work is licensed under a Creative Commons Attribution 4.0 International License.





