Manuscript accepted on : 07 February 2017
Published online on: --
Sepideh Rahimi¹ and Sahar Rahimi²
1Department of Plant Biotechnology,Payam Noor University (PNU), P.O.Box, 19395-3697 Tehran, Iran.
²Department of Food Science and Technology, Pharmaceutical Sciences branch Islamic Azad University, Tehran, Iran.
Corresponding Author E-mail: rahimisahar19@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2416
ABSTRACT: Aflatoxin is carcinogenic metabolites that produced by a number of species Aspergillus section Flavi. Cattle feed contaminated with Aflatoxin endanger human and animal health. The aim of this study is to determine amount of contamination to the Aspergillus section Flavi in five main animal feed with PCR molecular technique based on ITS regions and fungal colony. In this study, 121 ration samples were used from 21 industrial animal husbandry warehouses and silage in Tehran and Alborz Province. After isolating and culture in specific sphere of yeast extract sucrose agar (YESA), isolated Aspergillus fungi were studied by macroscopic and microscopic method. For molecular identification of the Aspergillus fungi from PCR method, ITS sequencing also was used and finally grown colonies were counted to determine an amount of the fungal contamination. The results showed that 67 samples from 121 ones were positive in which represents the 55/37% contamination amount to the Aspergillus fungi, and also the highest level of fungal contamination in imported barley, wheat bran, soybean meal and corn is about 14/16%, 12/5%, 10/83% and 10/83% respectively, while the lowest fungal contamination belongs to the internal barley with 7/5%. Therefore, there is the need to develop a simple, rapid and sensitive method for the detection of Aspergillus fungi. With regard to the high fungal contamination of imported cattle feed while its amount is very low in internal products, it is better that officials try to increase internal production as well as more controlling over the production, transport and import of cattle feed.
KEYWORDS: Aspergillus fungi; Aflatoxin; PCR; ITS; Cattle feed
Download this article as:| Copy the following to cite this article: Rahimi E, Rahimi S. Evaluation of the Cattle Ration feed Contamination Amount With the Aspergillus Fungi by PCR-Based Technique and Based on its Gene Sequences. Biosci Biotech Res Asia 2017;14(1). |
| Copy the following to cite this URL: Rahimi E, Rahimi S. Evaluation of the Cattle Ration feed Contamination Amount With the Aspergillus Fungi by PCR-Based Technique and Based on its Gene Sequences. Biosci Biotech Res Asia 2017;14(1). Available from: https://www.biotech-asia.org/?p=22602 |
Introduction
Mycotoxins are produced highly in cereal grains such as corn, sorghum, barley, wheat, cottonseed, peanut meal and fodder, before and during the harvest in humidity conditions (Richard et al. 2003). The Mycotoxin term first was introduced in 1962, after depth of one hundred thousand turkeys in Britain for an unknown disease. The disease caused by a toxin produced by the Aspergillus flavus fungi in birds feed (Wogan and Pong 1970). Following the crisis, scientists suspect other fungal metabolites may also be fatal. From 1960 to 1975 was the golden era of Mycotoxins studies, as scientists have done extensive research on these toxigenic factors (Bennett and Klich 2003). Mycotoxins are compounds produced by some strains of different fungal species especially Aspergillus, and also are dangerous for the health of humans and animals (Humans et al. 2002). Aspergillus genus has been classified in different categories based on morphological characteristics and genomic sequences which in the meantime Aspergillus section Flavi, Circumdati genus threatens public health by producing mycotoxins, particularly aflatoxin B1 which is known as the strongest natural hepatocarcinogen . They produce aflatoxin and can cause tissue necrosis, cirrhosis and liver cancer (Richard et al. 2003; Chu 1991). Strains of Aspergillus flavus, Aspergillus parasiticus and Aspergillus nomius are the most important species of this category (Yu et al. 2004; Varga et al. 2009; Ehrlich et al. 2003; Yu et al. 2000). Identification of Aspergillus fungi from various aspects such as pathogenicity, virulence and industrial is important in terms to whether be aflatoxin productive or unproductive (Bennett et al. 2007; Murphy et al. 2006). In order to distinguish these species, typically morphological characteristics are used such as macroscopic traits like color and colony growth rate, and also microscopic traits like morphology and measurement of Vesicle, Conidia, Conidiophore, Phialide and Metula (Klich 2002; Klich and Pitt 1988). Aspergillus species have very similarities to each other in order to morphological traits and on the other hand, this method is expensive, time-consuming and finally also does not have a high degree of accuracy. Therefore, molecular techniques, such as PCR are used to investigate the presence or absence of a gene, is faster and more accurate detection of aflatoxin-productive Aspergillus fungi. The Real-Time PCR is another method which is automated, rapid, repeatable and highly accurate technique to differ aflatoxin productive fungal samples from unproductive ones (Geisen 1996; Shapira et al. 1996; Färber et al. 1997; Sweeney et al. 2000; Criseo et al. 2001). These techniques are used as proprietary methods and the final diagnosis, and microarray technology will be of great worldwide importance in the near future.
The aim of this study is to isolate contaminated samples to Aspergillus fungi with morphological and microscopy characteristics, and PCR molecular technique based on ITS region. Then investigate the contamination of Aspergillus fungi in five main cattle feed rations in Tehran and Alborz province industrial animal husbandry.
Materials and Methods
Samples Collection
121 samples were collected randomly from 21 industrial animal husbandry warehouses and silos in Tehran and Alborz province, each in the amount of 250-500 grams, including corn (A), internal barley (B1), imported barley (B2), wheat bran (D) and soybean meal (E). Sampling time was 2014 – 2016 summer and winter respectively. These samples were placed in sterile bags and after encoding, (the name of animal husbandry, cattle feed type, date of collection) they were transferred to the laboratory. Samples were stored at -20°C until were been used.
Isolation Aspergillus fungi from Cattle feed Rations
First, for isolating Aspergillus fungi from cattle feed, 5 ml of distilled water was added to each cattle feed sample test tubes and after 2 hours, the supernatant liquid was collected; after vortex and centrifugation, 200 microliters of the mixture that obtained by sterile micropipette in saboro dextrose agar culture containing chloramphenicol, (to prevent the growth of bacteria and yeasts) was transferred; the plates was kept in the dark place at a temperature of 25 ° C for a week (Samson et al. 2004). Then grown isolated mixtures in each plate were examined macroscopically and microscopically (Klich and Pitt 1988). Colonies that showed Aspergillus characteristics, were purified and transferred to yeast extract sucrose agar (YESA) by using methods in which culturing in slope within the tube.
DNA Extraction
For DNA extraction, a few microliters of stored spore suspension of Aspergillus isolates was transferred to the plate containing YESA and the colony were used for DNA isolation for a week. 500 microliters of buffer (containing 1 M Tris-HCL (pH = 8), 0.5 M EDTA (pH = 8), 7.45 g KCl), 60 mg of fungal myceliums was added to Aspergillus colonies; they were crushed by vortex and manual method for 45 seconds and then centrifuged for 10 min with 5000 RPM. The supernatant liquid was transferred to a new tube and 300 microliters of cold isopropanol (which is kept at -20°C) was added; cell lysis and isopropanol were mixed by shaking micro-tubes, and then was centrifuged for 10 minutes with 12000 RPM. This time the supernatant liquid was removed and about 0.8 microliters of 70% alcohol added to the sediment and after 15 min we put it in the incubator at 37 ° C to evaporate remaining alcohol; finally, 50 microliters of deionized distilled water was added to the sediment and DNA dissolved with distilled water by gently tapping. The obtained liquid as pure DNA solution was kept in freezer at -20°C until required for use.
PCR Amplification
For molecular identification of cattle feed rations Aspergillus, ITS gene parts were used in this study and by using OLIGO7 powerful software, primers were designed based on standard sequences in the gene bank (Pryce et al. 2003). Proper application of the primers was determined by using the BLAST software. So based on these parts, ITS1-5.8-ITS2 general primers were made by Macrogene company. 5 microliters of extracted DNA (Table 1), 1 microliters of each of the Forward and Reverse primers, 10 microliters of master-mix PCR of Amplicon company [which contains 0.2 units per microliter of Taq DNA polymerase, 0.4 mM of each (dATP, dTTP, dCTP, dGTP) dNTP and 3 mM MgCl2] and the necessary amount of sterile deionized distilled water (ddH2O) was added to reach final volume of 20 microliters. In this study, the prepared master mix of Amplicon Company was used. Finally, the materials mixed slowly and were placed inside the thermo cycler device. All the steps were done under sterile conditions, laminar hood and within the ice dish. In the next step, for strengthen the gene in PCR reactions, genomic of isolated Aspergillus species was done with thermal model by using Forward, Reverse and DNA primers. PCR product with markers by bp100 molecular weight was electrophoresed on 1% Agarose gel containing Ethidium bromide, and it was studied by using documentation gel device. The PCR thermal program was performed according to Table 2.
Table 1: Sequences of the nucleotide primers used in this study
| Primer code | Target gene | Primer sequences | PCR product size (bp) | Accession no |
| ITS-1for | ITS | 5′- GGCTTTGTCACCCGCTCTGT -3′ | 691 | AF027863.1 |
| ITS-2rev | 5′- ACGACCATTATGCCAGCGTCC -3′ |
Table 2: Heat program used for PCR
| 1 cycle | 34 cycle | 1 cycle | ||||||||
| PCR steps |
Initial denaturation | Denaturation | Annealing | Extention | Final extention | |||||
| Tm | Time | Tm | Time | Tm | Time | Tm | Time | Tm | Time | |
| ITS | 95◦С | 2 min | 95◦С | 30sec | 62◦С | 45sec | 72◦С | 45sec | 72◦С | 7 min |
Results
Morphologic and Microscopic Characteristics of Aspergillus fungi in Cattle feed
A total of 121 samples of cattle feed collected from industrial animal husbandry were cultured and examined. After a week, Aspergillus species were isolated on YES (Samson et al. 2004). To do so, the form and color of the surface and back of colonies on plates, the structure of mycelium, conidiofor and phyalids, the form, size and color of spores and their accumulation were examined using mycological keys and reliable resources (Klich and Pitt 1988; Peterson 2003).
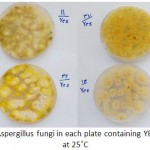 |
Figure 1: Grown isolates of Aspergillus fungi in each plate containing YESA were cultured for a week at 25˚C
|
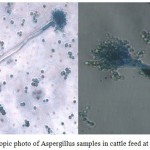 |
Figure 2: Microscopic photo of Aspergillus samples in cattle feed at 40x magnification
|
Molecular Identification of Isolated Aspergillus fungi by PCR Method
121 samples that collected by using the PCR method, were studied to identify Aspergillus fungi. After DNA extraction, the DNA concentration was measured by using NANO drop spectrophotometer. The amount of DNA was homogenized in all samples until get to 100 ngr. As noted above, 121 samples were randomly selected which is contained corn (A), internal barley (B1), imported barley (B2), wheat bran (D) and soybean meal (E). ITS general gene parts were used to isolate Aspergillus fungi in which the results of PCR products showed the 691 bp band development, and revealed that 67 of tested samples from 121 belongs to the Aspergillus genus which indicative 55.37% of Aspergillus fungi contamination. It is noteworthy that two standard samples (5004 Aspergillus flavus and 5018 Aspergillus parasiticus) were present at all stages for positive controlling (Figure 3 and Table 3).
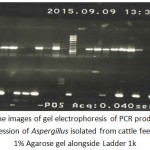 |
Figure 3: The images of gel electrophoresis of PCR products for ITS gene expression of Aspergillus isolated from cattle feed ration in 1% Agarose gel alongside Ladder 1k
|
Table 3: The PCR products of ITS genes are tested. Black box represents expressed gene and gray box denotes no expressed gene .(F) stands for the standard Aspergillus flavus and (P) stands for the standard Aspergillus parasiticus
| Sample Type |
Number | Isolate | ITS | Sample Type |
Number | Isolate | ITS | Sample Type |
Number | Isolate | ITS | Sample Type |
Number | Isolate | ITS |
| 1:A | 1 | _ | 5:E | 34 | A. flavus | + | 11:D | 67 | A. nomius | + | 17:E | 100 | _ | ||
| 1:B2 | 2 | A. flavus | + | 6:A | 35 | _ | 11:E | 68 | A. flavus | + | 18:A | 101 | A.parasiticus | + | |
| 1:B2 | 3 | A. flavus | + | 6:B1 | 36 | _ | 11:E | 69 | A. nidulans | + | 18:B2 | 102 | _ | ||
| 1:B2 | 4 | A. nomius | + | 6:D | 37 | A. nidulans | + | 12:A | 70 | _ | 18:D | 103 | _ | ||
| 1:B2 | 5 | _ | 6:E | 38 | A. flavus | + | 12:B1 | 71 | A. flavus | + | 18:E | 104 | _ | ||
| 1:B2 | 6 | A. flavus | + | 7:A | 39 | A. flavus | + | 12:B2 | 72 | _ | 19:A | 105 | _ | ||
| 1:C | 7 | _ | 7:A | 40 | A. flavus | + | 12:D | 73 | _ | 19:A | 106 | _ | |||
| 1:D | 8 | A. nidulans | + | 7:B1 | 41 | A. nidulans | + | 12:E | 74 | _ | 19:B2 | 107 | A. flavus | + | |
| 1:E | 9 | A. flavus | + | 7:B1 | 42 | _ | 13:A | 75 | _ | 19:B2 | 108 | A.parasiticus | + | ||
| 1:E | 10 | A. nomius | + | 7:D | 43 | A. flavus | + | 13:B1 | 76 | A. flavus | + | 19:B2 | 109 | _ | |
| 2:A | 11 | _ | 7:E | 44 | _ | 13:D | 77 | A. flavus | + | 19:D | 110 | A. flavus | + | ||
| 2:B1 | 12 | A.parasiticus | + | 8:A | 45 | _ | 13:E | 78 | _ | 19:E | 111 | _ | |||
| 2:B1 | 13 | A. flavus | + | 8:B1 | 46 | A. flavus | + | 14:A | 79 | A. flavus | + | 20:A | 112 | A. nidulans | + |
| 2:B2 | 14 | A. flavus | + | 8:D | 47 | A.parasiticus | + | 14:B1+C | 80 | A.parasiticus | + | 20:A | 113 | A. flavus | + |
| 2:D | 15 | _ | 8:E | 48 | _ | 14:D | 81 | _ | 20:A | 114 | A. flavus | + | |||
| 2:E | 16 | A. flavus | + | 9:A | 49 | A. flavus | + | 14:E | 82 | _ | 20:B2 | 115 | _ | ||
| 2:E | 17 | A. nidulans | + | 9:B2 | 50 | A. nidulans | + | 15:A | 83 | A. flavus | + | 20:D | 116 | A. flavus | + |
| 2:E | 18 | A. nidulans | + | 9:B2 | 51 | A. flavus | + | 15:A | 84 | _ | 20:E | 117 | A. nidulans | + | |
| 3:A | 19 | _ | 9:D | 52 | A. flavus | + | 15:B2 | 85 | A. nidulans | + | 21:A | 118 | _ | ||
| 3:B2 | 20 | A. flavus | + | 9:D | 53 | A. flavus | + | 15:B2 | 86 | A. flavus | + | 21:B2 | 119 | A.parasiticus | + |
| 3:D | 21 | _ | 9:D | 54 | _ | 15:D | 87 | A. flavus | + | 21:D | 120 | _ | |||
| 3:E | 22 | A. flavus | + | 9:D | 55 | A. flavus | + | 15:D | 88 | A.parasiticus | + | 21:E | 121 | _ | |
| 4:A | 23 | A. flavus | + | 9:E | 56 | A.parasiticus | + | 15:E | 89 | _ | F | ST | A. flavus | + | |
| 4:A | 24 | A. flavus | + | 10:A | 57 | _ | 16:A | 90 | _ | P | ST | A.parasiticus | + | ||
| 4:B2 | 25 | A.parasiticus | + | 10:A | 58 | A. nomius | + | 16:B1 | 91 | _ | |||||
| 4:D | 26 | A. nidulans | + | 10:B1 | 59 | A. flavus | + | 16:B2 | 92 | _ | |||||
| 4:D | 27 | _ | 10:B2 | 60 | _ | 16:C | 93 | _ | |||||||
| 4:E | 28 | _ | 10:B2 | 61 | A.parasiticus | + | 16:D | 94 | _ | ||||||
| 5:A | 29 | _ | 10:D | 62 | _ | 16:E | 95 | _ | |||||||
| 5:B2 | 30 | _ | 10:E | 63 | A. flavus | + | 17:A | 96 | _ | ||||||
| 5:B2 | 31 | A.parasiticus | + | 11:A | 64 | A. flavus | + | 17:B1 | 97 | A.parasiticus | + | ||||
| 5:B2 | 32 | A. flavus | + | 11:A | 65 | _ | 17:C | 98 | _ | ||||||
| 5:D | 33 | A. flavus | + | 11:B1+B2 | 66 | _ | 17:D | 99 | _ |
Counting the Fungal Colonies in Cattle feed Rations and Estimating the Amount of Contamination
The abundance of fungal contamination in corn samples of cattle feed was found when different analysis and isolation of different isolates were done; 29 fungal colonies out of grown corn isolated samples were grew up of which 13 colonies were related to Aspergillus. It can be said that the amount of corn contamination with Aspergillus is about 10.83 percent out of total grown isolates level. The percentage is our own expectations as well because corn fungal contamination is caused by being imported from tropical countries, transferring in hot and humid environment and improper storage conditions in shipping warehouses. Therefore, this issue requires special attention to internal production and also monitoring imports as well.
Also the abundance of fungal contamination in internal barley samples of cattle feed was found when different analysis and isolation of different isolates were done; 13 fungal colonies grew out of grown internal barley isolates sample of which 9 colonies were related to Aspergillus. Compared to the total amount of grown fungal isolates, it can be said that the internal barley contamination is about 7.5 percent out of total Aspergillus fungi contamination.
The abundance of fungal contamination in imported barley samples of cattle feed was found when different analysis and isolation of different isolates were done; 26 fungal colonies grew out of grown imported barley isolates sample of which 17 colonies were related to Aspergillus. Compared to the total amount of grown fungal isolates, it can be said that the imported barley contamination is about 14.16 percent out of total Aspergillus fungi contamination.
The abundance of fungal contamination in wheat bran samples of cattle feed was found when different analysis and isolation of different isolates were done; 26 fungal colonies grew out of grown wheat bran isolates sample of which 15 colonies were related to Aspergillus. Compared to the total amount of grown fungal isolates, it can be said that the wheat bran contamination is about 12.5 percent out of total Aspergillus fungi contamination.
Finally, the abundance of fungal contamination in soybean samples of cattle feed was found when different analysis and isolation of different isolates were done; 25 fungal colonies grew out of grown soybean isolates sample of which 13 colonies were related to Aspergillus. Compared to the total amount of grown fungal isolates, it can be said that the soybean contamination is about 10.83 percent out of total Aspergillus fungi contamination.
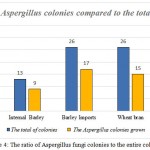 |
Figure 4: The ratio of Aspergillus fungi colonies to the entire colonies
|
 |
Figure 5: The amount of Aspergillus fungi contamination in cattle feed
|
Discussion
Aflatoxins are fungi secondary metabolites and mainly produce by Aspergillus section Flavi including Aspergillus flavus, Aspergillus parasiticus and Aspergillus nomius (Varga et al. 2009; Frisvad et al. 2005). Today, Mycotoxin productive fungal are world-wide distribution, and include more than 10 species of the genus Aspergillus that is often in Flavi section. The optimum temperature for producing aflatoxin has been reported between 24 -30 ° C (Ogundero 1987; Sorenson et al. 1967). In this study, we decided to survey the contamination of Aspergillus fungi in Iran through morphological and microscopic and molecular techniques such as PCR and growth colonies numeration (Gilbert and Vargas 2003). Macroscopic and microscopic studies of cattle feed rations showed that several species of Aspergillus have grew on these rations, and have contaminated them, that Aspergillus flavus, Aspergillus parasiticus were among them (Sales and Yoshizawa 2005). In the next phase, the PCR reaction was used for molecular detection of isolated Aspergillus of cattle feed . The universal ITS gene part was designed for this purpose and fungal associated with genus Aspergillus by 691 bp band was separated in the PCR reaction (Pryce et al. 2003; Rahimi et al. 2016a). The results of this study showed that based on the information, from 121 tested samples of cattle feed rations, 55.37 percent of all have contamination with the Aspergillus fungi (Scherm et al. 2005; RAHIMI et al. 2016b).
Sales et al (2005) studied 78 cattle feed samples from Thailand and Vietnam, and then they reported 94% contamination of samples by A. parasiticus and A. flavus species (Sales and Yoshizawa 2005). Halt also examined wheat, barley and corn used as cattle feed in Croatia and found that Aspergillus flavus is the main cause of contamination (Halt 1994). Scherm et al in 2005 introduced aflatoxin as a secondary metabolite which produced by Aspergillus section Flavi, especially Aspergillus flavus, Aspergillus parasiticus and Aspergillus nomius (Scherm et al. 2005). Sheila Okoth and colleagues (2012) studied 258 corn samples in Kenya by using the PCR method; they used genes aflQ and aflD aflatoxin productive gen to identify Aspergillus flavus and Aspergillus parasiticus toxin species, and attributed the maximum amount of contamination to Aspergillus section Flavi. They reported that the highest and the lowest levels are belonging to aflatoxins toxins B and G (Okoth et al. 2012). Villa and markesi in 2009 were investigated B1 aflatoxin contamination in 55 samples of wheat by using HPLC and immunoaffinity column. They declared that 3/56% of B1 aflatoxin was contaminated with amount of 1.42 ng/g (Villa and Markaki 2009).
In this study, the Aspergillus section Flavi fungi contamination of cattle feed were determined within the scope of contamination in food products which represents the accommodation of the results to previous researches. The results showed that the imported barley and wheat bran have the highest amount of Aspergillus contamination by 16/14% and 12.5% respectively, and the lowest was related to the internal barley with the 5/7%. According to the World Food and Agriculture Organization reports, FAO (Food and Drug Administration) annually 20 percent of food products are contaminated with fungal toxins in the world in which the contamination caused by aflatoxin has greater level (Van Egmond et al. 2007). The FAO announced that the maximum permitted level of aflatoxin in cattle feed ration should be 20 ppb (STEP 2004). The surveys conducted in this study indicates that the highest contamination rates of cattle feed is belong to imported feeds which should not exceed this limit, generally this issue is important because consumption of contaminated cattle feed with Aspergillus, which exposed humans and animals to aflatoxins as a threat to the health of humans and animals (Smith et al. 1995; Kabak et al. 2006).
Conclusion
Since studies show that the most contamination of cattle feed is belong to imported cattle feed, so with the better management and supervision on the quality of imports, aerated silo and barn of cattle feed rations, training how to keep cattle feed rations to farmers and experts can be partially prevented pathogenic fungi toxins and prevent of Mycotoxin entry to animal and human health cycle, and finally we can sample in large scale and in different provinces until the results can be generalized to the whole country.
References
- Bennett J, Kale S, Yu J (2007) Aflatoxins: background, toxicology, and molecular biology. In: Foodborne diseases. Springer, pp 355-373.
CrossRef - Bennett J, Klich M (2003) Mycotoxins. Clinical microbiology reviews 16 (3):497.
CrossRef - Chu FS (1991) Mycotoxins: food contamination, mechanism, carcinogenic potential and preventive measures. Mutation Research/Genetic Toxicology 259 (3-4):291-306.
CrossRef - Criseo G, Bagnara A, Bisignano G (2001) Differentiation of aflatoxin‐producing and non‐producing strains of Aspergillus flavus group. Letters in Applied Microbiology 33 (4):291-295.
CrossRef - Ehrlich KC, Montalbano BG, Cotty PJ (2003) Sequence comparison of aflR from different Aspergillus species provides evidence for variability in regulation of aflatoxin production. Fungal Genetics and Biology 38 (1):63-74.
CrossRef - Färber P, Geisen R, Holzapfel W (1997) Detection of aflatoxinogenic fungi in figs by a PCR reaction. International Journal of Food Microbiology 36 (2):215-220.
CrossRef - Frisvad JC, Skouboe P, Samson RA (2005) Taxonomic comparison of three different groups of aflatoxin producers and a new efficient producer of aflatoxin B 1, sterigmatocystin and 3-O-methylsterigmatocystin, Aspergillus rambellii sp. nov. Systematic and Applied Microbiology 28 (5):442-453.
CrossRef - Geisen R (1996) Multiplex polymerase chain reaction for the detection of potential aflatoxin and sterigmatocystin producing fungi. Systematic and Applied Microbiology 19 (3):388-392.
CrossRef - Gilbert J, Vargas EA (2003) Advances in sampling and analysis for aflatoxins in food and animal feed. Journal of Toxicology: Toxin Reviews 22 (2-3):381-422.
CrossRef - Halt M (1994) Aspergillus flavus and aflatoxin B1 in flour production. European journal of epidemiology 10 (5):555-558.
CrossRef - Humans IWGotEoCRt, Organization WH, Cancer IAfRo (2002) Some traditional herbal medicines, some mycotoxins, naphthalene and styrene. vol 82. World Health Organization,
- Kabak B, Dobson AD, Var Il (2006) Strategies to prevent mycotoxin contamination of food and animal feed: a review. Critical reviews in food science and nutrition 46 (8):593-619.
CrossRef - Klich M, Pitt J (1988) Differentiation of Aspergillus flavus from A. parasiticus and other closely related species. Transactions of the British Mycological Society 91 (1):99-108.
CrossRef - Klich MA (2002) Indentification of common Aspergillus species. Centraalbureau voor schimmelcultures,
CrossRef - Murphy PA, Hendrich S, Landgren C, Bryant CM (2006) Food mycotoxins: an update. Journal of food science 71 (5):R51-R65.
CrossRef - Ogundero VW (1987) Temperature and aflatoxin production by Aspergillus flavus and A. parasiticus strains from Nigerian groundnuts. Journal of basic microbiology 27 (9):511-514
- Okoth S, Nyongesa B, Ayugi V, Kang’ethe E, Korhonen H, Joutsjoki V (2012) Toxigenic potential of Aspergillus species occurring on maize kernels from two agro-ecological zones in Kenya. Toxins 4 (11):991-1007.
CrossRef - Peterson SW (2003) Identification of Common Aspergillus species by MA Klich (2002). Pp. 116. ISBN 90-70-351-46-3. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands. Price€ 28. Cambridge Univ Press,
- Pryce T, Palladino S, Kay I, Coombs G (2003) Rapid identification of fungi by sequencing the ITS1 and ITS2 regions using an automated capillary electrophoresis system. Medical mycology 41 (5):369-381.
CrossRef - Rahimi S, Sohrabi N, Ebrahimi MA, Tebyanian M, Rahimi S (2016a) Application of PCR in the Detection of Aflatoxinogenic and Non-aflatoxinogenic Strains of Aspergillus Flavus Group of Cattle Feed Isolated in Iran. Journal of Molecular Biology Research 6 (1):121.
CrossRef - Rahimi S, Sohrabi N, Ebrahimi Ma, Tebyanian M, Zadeh Mt, Rahimi S (2016B) Studying The Effect of Aflatoxin Genes Aflp And Aflq on Aspergillus Flavus And Aspergillus Parasiticus In The Cattle Feed Used In Industrial Animal Husbandries. Acta Medica 32:2091.
CrossRef - Richard J, Payne G, eds, Desjardins A, Maragos C, Norred W, Pestka J (2003) Mycotoxins: risks in plant, animal and human systems. CAST Task Force Report 139:101-103.
CrossRef - Sales AC, Yoshizawa T (2005) Updated profile of aflatoxin and Aspergillus section Flavi contamination in rice and its byproducts from the Philippines. Food additives and contaminants 22 (5):429-436.
CrossRef - Samson RA, Hoekstra ES, Frisvad JC (2004) Introduction to food-and airborne fungi. vol Ed. 7. Centraalbureau voor Schimmelcultures (CBS),
CrossRef - Scherm B, Palomba M, Serra D, Marcello A, Migheli Q (2005) Detection of transcripts of the aflatoxin genes aflD, aflO, and aflP by reverse transcription–polymerase chain reaction allows differentiation of aflatoxin-producing and non-producing isolates of Aspergillus flavus and Aspergillus parasiticus. International journal of food microbiology 98 (2):201-210.
CrossRef - Shapira R, Paster N, Eyal O, Menasherov M, Mett A, Salomon R (1996) Detection of aflatoxigenic molds in grains by PCR. Applied and Environmental Microbiology 62 (9):3270-3273.
CrossRef - Smith JE, Solomons G, Lewis C, Anderson JG (1995) Role of mycotoxins in human and animal nutrition and health. Natural toxins 3 (4):187-192.
CrossRef - Sorenson W, Hesseltine C, Shotwell OL (1967) Effect of temperature on production of aflatoxin on rice by Aspergillus flavus. Mycopathologia et mycologia applicata 33 (1):49-55.
CrossRef - STEP CA (2004) JOINT FAO/WHO FOOD STANDARDS PROGRAMME CODEX COMMITTEE ON FOOD ADDITIVES AND CONTAMINANTS Thirty-sixth Session Rotterdam, The Netherlands, 22-26 March 2004.
CrossRef - Sweeney MJ, Pàmies P, Dobson AD (2000) The use of reverse transcription-polymerase chain reaction (RT-PCR) for monitoring aflatoxin production in Aspergillus parasiticus 439. International Journal of Food Microbiology 56 (1):97-103.
CrossRef - Van Egmond HP, Schothorst RC, Jonker MA (2007) Regulations relating to mycotoxins in food. Analytical and bioanalytical chemistry 389 (1):147-157.
CrossRef - Varga J, Frisvad J, Samson R (2009) A reappraisal of fungi producing aflatoxins. World Mycotoxin Journal 2 (3):263-277.
CrossRef - Villa P, Markaki P (2009) Aflatoxin B 1 and ochratoxin A in breakfast cereals from Athens market: occurrence and risk assessment. Food Control 20 (5):455-461.
CrossRef - Wogan GN, Pong RS (1970) Aflatoxins. Annals of the New York Academy of Sciences 174 (2):623-635.
CrossRef - Yu J, Chang P-K, Ehrlich KC, Cary JW, Bhatnagar D, Cleveland TE, Payne GA, Linz JE, Woloshuk CP, Bennett JW (2004) Clustered pathway genes in aflatoxin biosynthesis. Applied and environmental microbiology 70 (3):1253-1262.
CrossRef - Yu J, Woloshuk CP, Bhatnagar D, Cleveland TE (2000) Cloning and characterization of avfA and omtB genes involved in aflatoxin biosynthesis in three Aspergillus species. Gene 248 (1):157-167.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





