How to Cite | Publication History | PlumX Article Matrix
Genetic Biodiversity analysis of two Mitochondrial genes in Arabian and Thoroughbred Horses
Karima F. Mahrous, Heba I. Shafey, Esraa A. Balabel and Othman E. Othman
Cell Biology Department, National Research Centre, Dokki, Egypt.
DOI : http://dx.doi.org/10.13005/bbra/2413
ABSTRACT: More than 300 horse breeds or types are present all over the world and employed in different activities. This work aimed to identify genetic variations and SNPs in two effective mtDNA genes; ATP6 and ND2; among Arabian and Thoroughbred horses.The primer used in this study amplified 340-bp fragments from ATP6 gene. The results showed the presence of 6 polymorphic sites leading to the construction of 7 different haplotypes which were submitted to GenBank database with the accession numbers KX377925-KX377931. For ND2 gene, the results showed the presence of 6 polymorphic sites leading to the construction of 5 different haplotypes. The sequences of detected haplotypes of horse ND2 gene were submitted to GenBank database with the accession numbers KX396591-KX396595. Neighbor-joining trees were constructed using 24 samples for ATP gene and 20 samples for ND2 gene with 10 reference sequences and the result declared the affinities of all tested animals to Equus caballus subspecies.In conclusion, the identification of genetic variations and SNPs in horse mitochondrial genes like ATP6 and ND2 genes are of great interest because they have highly significant effect and play important roles in different characteristics associated with speed and force which constitute race performance efficiency.
KEYWORDS: Arabian horses;ATP6 gene; Genetic biodiversity; ND2 gene; genetic biodiversity Thoroughbred horses;
Download this article as:| Copy the following to cite this article: Mahrous K. F, Shafey H. I, Balabel E. A, Othman O. E. Genetic Biodiversity analysis of two Mitochondrial genes in Arabian and Thoroughbred Horses. Biosci Biotech Res Asia 2017;14(1). |
| Copy the following to cite this URL: Mahrous K. F, Shafey H. I, Balabel E. A, Othman O. E. Genetic Biodiversity analysis of two Mitochondrial genes in Arabian and Thoroughbred Horses. Biosci Biotech Res Asia 2017;14(1). Available from: https://www.biotech-asia.org/?p=21865 |
Introduction
The Animal domestication played an important role in human civilization and changing of man’s lifestyle from hunter-gatherer to agricultural one.1 The horse (Equus ferus caballus L.) was firstly domesticated in the Eurasian region.2 More than 300 horse breeds – or actually types because sometimes the use of breed term for horses is not right – are present all over the world and employed in different activities.3
The Arabian horse is a breed originated on the Arabian Peninsula with a distinctive head shape and high tail carriage. Arabian horse is one of the most easily recognizable horse breeds in the world and also it is one of the oldest breeds with archaeological evidence of horses in the Middle East.4 Arabian horses have spread around the world by both war and trade, used to improve other breeds by adding speed, refinement, endurance, and strong bone.5 Today, Arabian bloodlines are found in almost every modern breed of riding. The Arabian developed in a desert climate and was prized by the nomadic Bedouin people. It is a versatile breed; Arabian horses dominate the discipline of endurance riding and compete today in many other fields of equestrian sport.6 They are one of the top ten most popular horse breeds in the world. They are now found worldwide, including the United States and Canada, United Kingdom, Australia, continental Europe, South America (especially Brazil) in addition to their land of origin, the Middle East horse.7
The Thoroughbred is a horse breed known for its use in horse racing. It is developed in at 17thcentury in England, when native mares were crossbred with Oriental stallions of Arabian, Barb and Turkoman breeding (www.britannica.com/animal/Thoroughbred). During the 18thand 19thcenturies, the Thoroughbred breed spread throughout the world; they were imported into North America, Australia, Europe, Japan and South America (www.ansi.okstate.edu/breeds/horses/thoroughbred). Thoroughbreds are used mainly for racing, but are also bred for other riding disciplines such as show jumping, combined training, dressage, polo and fox hunting. They are also commonly crossbred to create new breeds or to improve existing ones, and have been influential in the creation of the Quarter Horse, Standardbred, Anglo-Arabianand other various horse breeds (www.tbheritage.com).
The race efficiency characteristics of horses are inherited mainly through the maternal genetics with highly significant effect.8 MtDNA genes plays an important role in various characteristic associated with speed and force which have effect on race performance efficiency.9 The variations in mtDNA genes influence the potential and stamina of different horse breeds.10,3 This work aimed to identify genetic variations and SNPs in two effective mtDNA genes; ATP6 and ND2; among Arabian and Thoroughbred horses.
Material and Methods
Blood Samples and Genomic DNA Extraction
Blood samples were collected from 24 horses belonging to two breeds; Arabian and Thoroughbred horses. Genomic DNA was extracted from the whole blood according to the method described by11 with minor modifications. Briefly, Blood samples were mixed with cold 2x sucrose-triton and centrifuged at 5000 rpm for 15 min at 4°C. The nuclear pellet was suspended in lysis buffer, sodium dodecyl sulfate and proteinase K and incubated overnight in a shaking water bath at 37°C. Nucleic acids were extracted with saturated NaCl solution. The DNA was picked up and washed in 70% ethanol. The DNA was dissolved in 1xTE buffer. The DNA concentration was determined, using Nano Drop1000 thermo scientific spectrophotometer and then diluted to the working concentration of 50 ng/μl.
Polymerase Chain Reaction (PCR)
The PCR amplifications were conducted in a 50 μL volume containing 5 μL of 10x reaction buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.2 μM each primer (Table 1), 1.5U Taq DNA polymerase (Fermentas, Germany) and approximately 100 ng genomic DNA. The reaction was cycled for 1 min at 94°C, 1 min at an optimized annealing temperature that was determined for each primer (Table 1) and 2 min at 72°C for 35 cycles. The PCR products were electrophoresed on 2% agarose gel stained with ethidium bromide to test the amplification success. The PCR products were purified and sequenced by Macrogen Incorporation (Seoul, Korea).
Table 1: Sequences of primers used in this study and amplification conditions
| Gene | Primer sequences
5΄———–3΄ |
PCR conditions | Size of amplified fragments |
|
ATP6 |
CTA TGG GCA GGG ACA GTA TT
AAA GGC TTA CCA GGA GAG TG |
95°C 1 min
58°C 1 min 72°C 1 min |
340-bp |
|
ND2
|
CCC CGA ACC ATA GAA GCC TC
AGA CCG CCT CAG CCT CCT AC |
94°C 1 min
57°C 1 min 72°C 1 min |
362-bp |
Data Analysis
The sequences of amplified fragments of each tested genes were aligned using the BioEdit software.12 The DnaSP 5.00 software13 was used to identify the polymorphic sites and SNPs between different animals. Neighbor-joining tree was computed with Mega version 5.0.14
Results and Discussion
The mitochondrial genome is a principal component in the metabolism process of living organism. The mitochondrion is responsible for the producing of about 95% energy for eukaryotic cells and it encodes 13 proteins through four protein complexes. These protein groups are cytochrome bcl, cytochrome oxidase, nicotinamide adenine dinucleotide (NADH) dehydrogenase and adenosine triphosphate (ATP) synthase. These proteins are involved in the oxidative phosphorylation process which has an essential role in energy metabolism.15 Also in the mitochondrion, there are seventy-six nuclear genes encoding proteins which are functional in oxidative phosphorylation system.16 All these proteins can modify the metabolic fitness by functional interactions and have an important role in race efficiency characteristics of horses like force and speed.17-19
Many studies focused on mtDNA genes and their effective roles in racing and sporting efficiency of horses as well as fitness and performance phenotypes.8,20 Other studies discussed the effect of the combination and interaction between mitochondrial genes on the efficiency of race horses. So, the genetic characterization and phylogeny of mitochondrial genes will be of great interest in horses due to their important effects on horse race performances. This study aimed to identify the genetic variations and SNPs of two effective mtDNA genes; ATP6 and ND2; in Arabian and Thoroughbred horses. This work also aimed to clarify the phylogeny relationship between these two breeds.
MT-ATP6 gene
ATP6 is a mitochondrial gene encoding the MT-ATP6 protein which forms one part of a large enzyme called ATP synthase. This enzyme is responsible for the final step of oxidative phosphorylation process. ATP has an important role in the energy transferring within the cell, diffusing from the place in which it is produced to the place in which it is utilized. Thirty percent of total ATP consumption is utilized in ion transport processes.21 Due to the important role of ATP6 in energy process which has great effect on the race efficiency of horses, its genetic diversity in Arabian and Thoroughbred horses was included in this work.
The primer used in this study amplified 340-bp fragments from ATP6 gene in Arabian and Thoroughbred horses. These amplified fragments were sequenced and the nucleotide sequences of 24 samples belonging to these two horse breeds were aligned using BioEdit software. DnaSP 5.00 software was used to identify the sequence variation and polymorphic sites in the aligned sequences.
The results showed the presence of 6 polymorphic sites leading to the construction of 7 different haplotypes (Fig. 1). The sequences of all detected haplotypes of ATP6 were submitted to GenBank database with the accession numbers KX377925-KX377931. The 14 Arabian samples showed 6 haplotypes with 6 polymorphic sites whereas the 10 Thoroughbred horses showed 4 haplotypes with 4 polymorphic sites. Haplotype no. 6 was the most detected oneand found in 6 samples; 3 horses samples from each breed followed by haplotype no. 5 which was found in 5 samples; 4 from Thoroughbred and one from Arabian horses. Haplotypes nos. 2 an6 6 are specific for Arabian breed which were present in 3 and 4 samples, respectively whereas haplotype no. 7 is specific for Thoroughbred and present in 2 samples. Fig. 2 declared the presence of 7 clusters; 3 specific for Arabian horses, 1 specific for Thoroughbred and 3 mixing for both.
 |
Figure 1: The sequences of seven detected haplotypes of ATP6 gene with 6 polymorphic sites in red
|
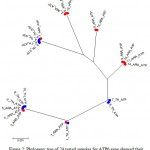 |
Figure 2: Phylogeny tree of 24 tested samples for ATP6 gene showed their segregation in 7 clusters.
|
ARA: Arabian in red,
TH: Thoroughbred in blue
The haplotype diversities (HD) in Arabian and Thoroughbred horses were 0.857 and 0.778 and the average nos. of nucleotide diversities (K) were 2.363 and 2.067, respectively. The HD and K in all 24 tested samples were 0.866 and 2.326, respectively. The nucleotide diversity was 0.00695 for Arabian, 0.00608 for Thoroughbred and 0.00684 for all tested horses (Table 2).The statistical analysis of haplotypes diversity of Arabian and Thoroughbred horses showed that the average number of nucleotide differences between two populations was 2.386, the average number of nucleotide substitutions per site between populations (Dxy) was 0.00702 and the number of net nucleotide substitutions per site between populations (Da) was 0.00050.
Table 2: The genetic diversity data of ATP6 gene
| Breed | Arabian | Thoroughbred | Total |
| No. of samples | 14 | 10 | 24 |
| No. of polymorphic sites (S) | 6 | 4 | 6 |
| No. of haplotypes (H) | 6 | 4 | 7 |
| Haplotype diversity (HD) | 0.857 | 0.778 | 0.866 |
| Average No. of nucleotide differences (K) | 2.363 | 2.067 | 2.326 |
| Nucleotide diversity (p) | 0.00695 | 0.00608 | 0.00684 |
Neighbor-joining (Phylogeny) tree was constructed using the Mega 5.0 software. The sequences of the 24 tested samples were aligned with 10 sequences of different breeds and isolates of horses around world. The reference sequences used were: NC_001640.1, EU939445.3, EF597512.1, EF597513.1, EF597514.1, F038160.1, X79547.1, AP013078.1, X97337.1 and JX312729.1. The results showed that all 24 tested horses belonging to Arabian and Thoroughbred horses are grouped with 8 out of 10 references which are Equus caballus and separated from the other two references; JX312729.1 (Equus burchellii) and X97337.1 (Equus asinus) (Fig. 3). This result declared that all tested Arabian and Thoroughbred horses are belonging to Equus caballus subspecies.
 |
Figure 3: Phylogeny tree of 24 tested samples for ATP6 gene with ten reference sequences.
|
ARA: Arabian in red,
TH: Thoroughbred in blue and
KF038160, EF597514, EU939445, AP013078, EF597513, X79547, NC_001640, EF597512, X97337 and JX312729: accession number of references in green
MT-ND2 gene
ND2 gene is a mitochondrial gene encoding NADH dehydrogenase subunit 2 protein.21 MT-ND2 is located in mitochondrial DNA and produces a 39 kDa protein composed of 347 amino acids.22 MT-ND2 is a subunit of the respiratory chain complex which belongs to the minimal assembly for core protein required to catalyze NADH dehydrogenase.23 The identification of genetic variations in this gene through different horse breeds has a great interest due to its role in the physiological characteristics associated with race performance in horses.
The primer used in this study amplified 362-bp fragments from ND2 gene in Arabian and Thoroughbred horses. These amplified fragments were sequenced and the nucleotide sequences of 20 samples belonging to these two horse breeds were aligned to identify the sequence variation and polymorphic sites in the aligned sequences.
The results showed the presence of 6 polymorphic sites leading to the construction of 5 different haplotypes (Fig. 4). The sequences of detected haplotypes of horse ND2 gene were submitted to GenBank database with the accession numbers KX396591-KX396595. The six Arabian samples showed 2 haplotypes with one polymorphic site whereas the 14 Thoroughbred horses showed 4 haplotypes with 5 polymorphic sites. Fig. 5 declared the presence of a major cluster containing most of the tested samples; 4 from Arabian and 11 from Thoroughbred horses. The rest of tested samples are belonged to 4 small clusters; one is specific for Arabian horses (2 samples) and the other three clusters are specific for Thoroughbred horses containing one sample for each.
 |
Figure 4: The sequences of five detected haplotypes of ND2 genes with 6 polymorphic sites in red
|
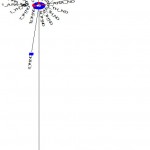 |
Figure 5: Phylogeny tree of 20 tested samples for ND2 gene showed their segregation in 5 clusters.
|
ARA: Arabian in red,
TH: Thoroughbred in blue
The haplotype diversities (HD) in Arabian and Thoroughbred horses were 0.5333 and 0.3956 and the average nos. of nucleotide diversities (K) were 0.533 and 0.714, respectively. The HD and K in all 20 tested samples were 0.4421 and 0.689, respectively. The nucleotide diversity was 0.00147 for Arabian, 0.00197 for Thoroughbred and 0.0019 for all tested horses (Table 3). The statistical analysis of haplotypes diversity of Arabian and Thoroughbred horses showed that the average number of nucleotide differences between two populations was 0.96, the average number of nucleotide substitutions per site between populations (Dxy) was 0.00191 and the number of net nucleotide substitutions per site between populations (Da) was 0.00018.
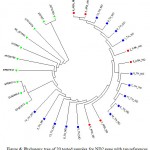 |
Figure 6: Phylogeny tree of 20 tested samples for ND2 gene with ten references.
|
ARA: Arabian in red,
TH: Thoroughbred in blue and
KF038160, EF597514, EU939445, AP013078, EF597513, X79547, NC_001640, EF597512, X97337 and JX312729: accession number of references in green
Table 3: The genetic diversity data of ND2 gene
| Breed | Arabian | Thoroughbred | Total |
| No. of samples | 6 | 14 | 20 |
| No. of polymorphic sites (S) | 1 | 5 | 6 |
| No. of haplotypes (H) | 2 | 4 | 5 |
| Haplotype diversity (HD) | 0.5333 | 0.3956 | 0.4421 |
| Average No. of nucleotide differences (K) | 0.533 | 0.714 | 0.689 |
| Nucleotide diversity (p) | 0.00147 | 0.00197 | 0.0019 |
Neighbor-joining (Phylogeny) tree was constructed using the Mega 5.0 software. The sequences of the 20 tested samples of ND2 gene were aligned with 10 sequences of different breeds and isolates of horses around world which were mentioned above. The results – confirmed the findings of ATP6 gene – where all tested horses belonging to Arabian and Thoroughbred horses are grouped with 8 out of 10 references which are Equus caballus and separated from the other two references; JX312729.1 (Equus burchellii) and X97337.1 (Equus asinus) (Fig. 6).
In conclusion, the identification of genetic variations and SNPs in horse mitochondrial genes like ATP6 and ND2 genes are of great interest because they have highly significant effect and play important roles in different characteristics associated with speed and force which constitute race performance efficiency. ATP6 gene is considered more informative marker that ND2 for genetic biodiversity and affinities of different horse breeds.
References
- Zeder M. A. Domestication and early agriculture in the Mediterranean Basin Origins diffusion and impact. Proceedings of the National Academy of Sciences. 2008;105(33):11597-11604.
Cross Ref - Outram A. K., Stear N. A., Bendrey R., Olsen S., Kasparov A., Zaibert V., Thorpe N., Evershed R. P. The earliest horse harnessing and milking. Science. 2009;323(5919):1332-5.
Cross Ref - Ling Y., Ma Y., Guan W., Cheng Y., Wang Y., Han J., Jin D., Mang L., Mahmut H. Identification of Y chromosome genetic variations in Chinese indigenous horse breeds. J Hered. 2010;101(5):639-43.
Cross Ref - Głażewska I. Speculations on the origin of the Arabian horse breed. Livestock Science. 2010;129(1–3):49-55.
Cross Ref - Bowling A. T., Ruvinsky A. The Genetics of the Horse. 2000. CABI.
Cross Ref - Bower M.A., Campana M. G. Whitten M.,Edwards C. J., Jones H., Barrett E., Cassidy R.. Nisbet R. E. R.. Hill E. W., How C. J., Binns M. The cosmopolitan maternal heritage of the Thoroughbred racehorse breed shows a significant contribution from British and Irish native mares. Biology Letters. 2011;7(2):316-320.
Cross Ref - Zechner P., Sölkner J., Bodo I., Druml T., Baumung R., Achmann R., Marti E., Habe F., Brem G. Analysis of diversity and population structure in the Lipizzan horse breed based on pedigree information. Livestock Production Science. 2002;77(2–3):137-146.
Cross Ref - Harrison S. P., Turrion-Gomez J. L. Mitochondrial DNA: An important female contribution to thoroughbred racehorse performance. Mitochondrion. 2006;6(2):53-66.
Cross Ref - Lesage R., Simoneau J. A., Jobin J., Leblanc J., Bouchard C. Familial resemblance in maximal heart rate blood lactate and aerobic power. Hum Hered. 1985;35(3):182-9.
Cross Ref - Lindgren G., Backstrom N., Swinburne J., Hellborg L., Einarsson A., Sandberg K., Cothran G., Vila C., Binns M., Ellegren H. Limited number of patrilines in horse domestication. Nat Genet. 2004;36(4):335-6.
Cross Ref - Miller S. A., Dykes D. D., Polesky H. F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research. 1988;16(3):1215.
Cross Ref - Hall T. A. Bio Edit a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95-98.
- Librado P., Rozas J. DnaSP v5 a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451-2.
Cross Ref - Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA 5 molecular evolutionary genetics analysis using maximum likelihood evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731-9.
Cross Ref - Lopez-Barneo J., Pardal R., Ortega-Saenz P.Cellular mechanism of oxygen sensing. Annu Rev Physiol. 2001;63:259-87.
Cross Ref - Grossman L. I., Wildman D. E., Schmidt T. R., Goodman M. Accelerated evolution of the electron transport chain in anthropoid primates. Trends Genet. 2004;20(11):578-85.
Cross Ref - Blier P. U., Dufresne F., Burton R. S. Natural selection and the evolution of mtDNA-encoded peptides: evidence for intergenomic co-adaptation. Trends Genet. 2001;17(7):400-6.
Cross Ref - Rand D. M., Haney R. A., Fry A. J. Cytonuclear coevolution the genomics of cooperation. Trends Ecol Evol. 2004;19(12):645-53.
Cross Ref - da Fonseca R. R., Johnson W. E., O’Brien S. J., Ramos M. J., Antunes A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genomics. 2008;9:119.
Cross Ref - Barrey E. Reviewe Genetics and genomics in equine exercise physiology an overview of the new applications of molecular biology as positive and negative markers of performance and health. Equine Vet J Suppl. 2010;(38):561-8.
Cross Ref - Torroni A., Achilli A., Macaulay V., Richards M., Bandelt H. J. Harvesting the fruit of the human mt DNA tree. Trends in Genetics. 2006;22(6):339-345.
Cross Ref - Zong N. C., Li H., Li H., Lam M. P., Jimenez R. C., Kim C. S., Deng N., Kim A. K., Choi J. H., Zelaya I., Liem D., Meyer D., Odeberg J., Fang C., Lu H. J., Xu T., Weiss J., Duan H., Uhlen M., Yates J. R., Apweiler R., Ge J., Hermjakob H., Ping P. Integration of cardiac proteome biology and medicine by a specialized knowledge base. Circ Res. 2013;113(9):1043-53.
Cross Ref - Attardi G., Chomyn A., Doolittle R. F., Mariottini P., Ragan C. I. Seven unidentified reading frames of human mitochondrial DNA encode subunits of the respiratory chain NADH dehydrogenase. Cold Spring Harb Symp Quant Biol.1986;51(1):103-14.
Cross Ref

This work is licensed under a Creative Commons Attribution 4.0 International License.





