How to Cite | Publication History | PlumX Article Matrix
Hussein O. M. Al-Dahmoshi, Ayad M. J. Almamoori and Noor S. K. Al-Khafaji
Department of Biology- College of science, University of Babylon, Iraq.
Corresponding Author E-mail: dr.dahmoshi83@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2420
ABSTRACT: Copper is a unique heavy metal with good antibacterial properties. It is functionality associated with various mechanisms, including damaging the microbial DNA, altering bacterial protein synthesis and altering membrane integrity. The resistance of E. coli to cooper add another virulence trait that make it resistance to another heavy metals like silver and cadmium and resistance to many antibiotics which refluxed by same manner. Minimum inhibitory concertration (MIC) measurement were performed according to protocol of CLSI 2016. PCR were used to investigate presence of cusC gene using specify primer. The results of phenotypic investigation of copper resistance (tolerance) revealed that 16 (59.25%) of UPEC isolated have MIC ˂= 500 ppm while the rest 11(40.75%) have MIC>=1500 ppm. PCR results revealed that absence of cusC gene in all UPEC isolated (16 (59.25%) that have MIC ˂= 500 ppm while it presence in all those have MIC>=1500 ppm (except U5 and U8). As a conclusion this study state that the CusCFBA efflux system is the main copper extruding that can protect E. coli from oxidative damage by cooper.
KEYWORDS: UPEC; cooper/silver efflux; cusC; CusCFBA
Download this article as:| Copy the following to cite this article: Al-Dahmoshi H. O. M, Almamoori A. M. J, Al-Khafaji N. S. K. Investigation of Cuscfba Pump Among Uropathogenic Escherichia Coli (UPEC) Isolated from Women with Cystitis, Iraq. Biosci Biotech Res Asia 2017;14(1). |
| Copy the following to cite this URL: Al-Dahmoshi H. O. M, Almamoori A. M. J, Al-Khafaji N. S. K. Investigation of Cuscfba Pump Among Uropathogenic Escherichia Coli (UPEC) Isolated from Women with Cystitis, Iraq. Biosci Biotech Res Asia 2017;14(1). Available from: https://www.biotech-asia.org/?p=20566 |
Introduction
Uropathogenic Escherichia coli(UPEC) is the foremost causative agents of urinary tract infections (UTIs). UPEC are able to quickly invade, survive and multiply within the host cells and tissues of the urinary tract (Lüthje and Brauner, 2014; Abdulla et al., 2016). About 60% of women in the United States will have at least one UTI during their lifetime (especially cystitis). Symptoms of an uncomplicated bladder infection (cystitis) can be relatively mild, including frequent or urgent voiding and suprapubic pain (Bent et al., 2002; Colgan et al., 2007). The classical virulence factors were extensively studied while the resistance to metals like copper and silver were studied with less details. Heavy metals such as silver and copper are considered toxic to bacteria and can be used in industry of medical disposable like silver compounds which are used to coat urinary catheters in order to prevent nosocomial UTIs. UPEC have a set of genes that confer resistance to many heavy metals and especially copper and silver (Gupta et al., 2001; Johnson et al., 2006). Copper and silver have an antibacterial (bactericidal) effects and the resistant to them regard as a virulence traits. Copper antibacterial functionality is associated with various mechanisms, including damaging the microbial DNA, altering bacterial protein synthesis and altering membrane integrity (Warnes et al., 2010; Grass et al., 2011; Chaturvedi and Henderson, 2014). The mechanism of antibacterial action of silver depends on their reaction with thiol group in vital enzymes and inactivates them or interacts with DNA, resulting in marked enhancement of pyrimidine dimerization by photodynamic reaction and possible prevention of DNA replication and also generation of toxic free-radicals (Feng et al., 2000; Kim et al., 2007; Chen and Schluesener, 2008; Prabhu and Poulose, 2012).
Generally E. coli, have three systems that confer resesitance or tolerance to copper ions. The first of these, CopA, is an inner membrane P-type ATPase that actively pumps excess copper out of the cytoplasm. The second system consists of the multicopper oxidase CueO, which may protect periplasmic enzymes from copper-mediated damage. Finally, the Cus system is responsible for copper and silver resistance in E.coli (Franke et al., 2003; Rensing and Grass, 2003). Escherichia coli, frequently utilize CusCFBA efflux complexes in the resistance-nodulation-cell division (RND) family to expel diverse toxic compounds from the cell. cusCFBA operon dependent on the concentration of copper and silver ions within the cell. Within the cusCFBA operon CusA encodes an inner membrane protein belonging to the resistance-nodulation-division (RND) family. Phylogenetic analysis shows that CusA belongs to a subgroup of RND proteins involved in heavy metal extrusion (Nies, 2003). CusB is a membrane fusion/adaptor protein (MFP) and CusC is an outer membrane (OM) protein belonging to the outer membrane factor (OMF family). Together, CusABC likely forms a protein complex spanning the inner membrane, periplasmic space and the outer membrane, analogous to the well-studied AcrAB-TolC multidrug efflux pump from E. coli. CusF is a small periplasmic protein that binds copper and silver and may subsequently transport these ions to CusB. Thus, the CusCFBA proteins form a tetrapartite system dedicated to the efflux of heavy metal ions from E. coli (Kittleson et al., 2006; Loftin et al., 2007; Bagai et al., 2008). The current study aims to investigate copper resistance (tolerance) phenotypically and presence of the outer membrane part, cusC, as an evidence for presence of cusCFBA efflux pump.
Materials and Methods
Bacterial Isolates
Twenty seven UPEC isolated and diagnosed previously (By Viteck2 compact system/Biomeruex), were taken from Advance Microbiology laboratory at college of science-university of Babylon, Iraq. All isolates were recovered from women with cystitis.
Determination of Minimum Inhibitory Concentration of Copper
Agar dilution method was used to determine the minimum inhibitory concentration (MIC) of copper against 27 isolates of UPEC according to CLSI 2016. Briefly Muller-Hinton agar plates were supplemented with one of the 20 concentration of copper sulfate pentahydrate (CuSO4x5H2O) (Scharlab/Spain). The concertrations were 100, 200, 300, 400, 500, 600, 700, 800, 900, 1000, 1100, 1200, 1300, 1400, 1500, 1600, 1700, 1800, 1900, 2000 ppm. A suspension of UPEC isolates were calibrated with 0.5 McFarland and then 100 μl inoculated on Muller-Hinton agar supplemented with copper sulfate pentahydrate and incubated for 24 h at 37°C. The results of now growth were recorded as resistance and the least concentration of copper sulfate pentahydrate at which now growth appeared were recorder as MIC for copper (Reyes-Jara et al., 2016).
DNA Extraction
Genomic DNA of UPEC were extracted using FavorPrep Genomic DNA Mini Kit (Blood/Cultured Cell) according to instructions of manufactured company (Favorgen/Tiwan).
Primer Design, PCR and Gel Electrophoresis
The primer pairs used in this study were designed using primer 3 at Biology Workbench 3.2 (http://workbench.sdsc.edu). The designed primer pair then checked with primer-blast: (https://www.ncbi.nlm.nih.gov/tools/primerblast). The primer pairs sequence were cusC forward:5′-CGCCTTTAAAGAAGTGGCAG-3′ and cusC reverse:5′-CTGACGGGCATAATTCAGGT-3′. The PCR protocol were designed using Optimase Protocol Writer™. The 20 μl final volume of PCR reaction were mixed in 0.2 thin wall PCR tube containing 5 μl of premix, 5 μl of DNA, 3 μl of each of forward and reverse primer and the 4 μl of nuclease free water (Al-Alaq et al., 2016). The PCR conditions were 95°C for 2 minutes, 30 cycles of 95°C for 30 seconds, 58°C for 30 seconds, 72°C for 40 seconds. The final extension were 72°C for 5 minutes. All tubes were place in Prime thermocycler (Techno/UK). The PCR products were electrophoresed using 1.5% agarose gel (Condalab/Spain) stained with SimplySafe, using 100 bp DNA ladder (Eurex/Poland) and visualized using Quantum ST5 gel doc (Vilber/France).
Results and Discussion
The results of phenotypic investigation of copper resistance (tolerance) revealed that 16 (59.25%) of UPEC isolated have MIC ˂= 500 ppm while the rest 11(40.75%) have MIC>=1500 ppm (Table 1). Our data disagree with those gathered by Reyes-Jara et al. (2016) who found that 93% of isolates were inhibited with less than 500 ppm of copper. Generally different studies reported different MIC values of copper to E. coli like Sabry et al. (1997) who found that the level of copper resistance was reported to be 2.5 mM. while the copper MIC for Escherichia coli was 10 mM as stated by Roane and Kellogg (1995). Koowatananukul et al. (2010) found that most E. coli (92.2%) had an MIC of 6.4 mM to copper sulfate.
Table 1: Distribution of MIC for copper among UPEC isolates.
| UPEC isolates | Growth at | |||||
| 300 ppm | 400 ppm | 500 ppm | 1500 ppm | 1600 ppm | 1700 ppm | |
| U1 | + | + | + | + | – | – |
| U2 | + | + | + | + | + | + |
| U3 | + | + | + | + | + | + |
| U4 | + | + | + | + | – | – |
| U5 | + | + | + | + | – | – |
| U6 | + | + | + | + | + | + |
| U7 | + | + | + | + | + | + |
| U8 | + | + | + | + | + | + |
| U9 | + | + | + | + | – | – |
| U10 | + | + | + | + | – | – |
| U11 | + | + | + | + | + | + |
| U12 | + | – | – | – | – | – |
| U13 | + | – | – | – | – | – |
| U14 | + | + | – | – | – | – |
| U15 | + | + | – | – | – | – |
| U16 | + | – | – | – | – | – |
| U17 | + | – | – | – | – | – |
| U18 | + | + | – | – | – | – |
| U19 | + | – | – | – | – | – |
| U20 | + | + | – | – | – | – |
| U21 | + | – | – | – | – | – |
| U22 | + | – | – | – | – | – |
| U23 | + | + | – | – | – | – |
| U24 | + | + | – | – | – | – |
| U25 | + | + | – | – | – | – |
| U26 | + | – | – | – | – | – |
| U27 | + | + | – | – | – | – |
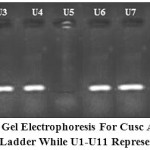 |
Figure 1: 1.5% Agarose gel electrophoresis for cusC amplicon (217 bp). Lane M 100 bp DNA Ladder while U1-U11 represent UPEC isolates.
|
PCR results revealed that absence of cusC gene in all UPEC isolated (16 (59.25%) that have MIC ˂= 500 ppm while it presence in all those have MIC>=1500 ppm (except U5 and U8) (figure 1). Although it have high MIC but U5 and U8 isolates have no cusC and may have another efflux system like CopA or CueO (Franke et al., 2001; Grass and Rensing, 2001; Macomber et al., 2007; Pontel and Soncini, 2009). Our results in accordance with Macomber et al. (2007) who found that E. coli protect it selves from copper-mediated oxidative stress prevent accumulation of significant intracellular concentrations of free copper ions by efflux.
As a conclusion this study state that the CusCFBA efflux system is the main copper extruding that can protect E. coli from oxidative damage by cooper.
References
- Abdulla, A.A., Al-Dahmoshi, H.O.M., Abed, T.A. and Muttaleb, W.H. (2016). Characterization of Multidrug Resistant Carbapenemases-Producing Escherichia coli and Klebsiella pneumoniae Isolates from Urinary Tract Infection. Journal of Chemical and Pharmaceutical Sciences, 9(3): 1116-1120.
- Al-Alaq, F.T., Abdulazeem, L., Al-Dahmoshi, H.O.M., Al-Khafaji, N.S. and Al-Wesawei, Y.A., (2016). PCR-based investigation of oxygenase among crude oil degrading bacteria in Hilla city, Iraq. International Journal of Pharm Tech Research, 9(5), pp.284-291.
- Bagai, I., Rensing, C., Blackburn, N. and McEvoy, M. (2008). Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry., 47(44), pp. 11408–14.
CrossRef - Bent, S., Nallamothu, B., Simel, D., Fihn, S. and Saint, S. (2002). Does this woman have an acute uncomplicated urinary tract infection?. JAMA. 287(20), pp. 2701–10.
CrossRef - Bower, J.M., Eto, D.S., Mulvey, M.A. (2005). Covert operations of Uropathogenic Escherichia coli within the urinary tract. Traffic 6(1):18-31.
CrossRef - Chaturvedi, K.S., Henderson, J.P.(2014).Pathogenic adaptations to host-derived antibacterial copper. Front. Cell. Infect. Microbiol. 4:3.
CrossRef - Chen, X. and Schluesener, H.J. (2008). Nanosilver: A nanoproduct in medical application. Toxicology Letters, 176(1), pp. 1–12.
CrossRef - Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. CLSI supplement M100S. Wayne, PA: Clinical and Laboratory Standards Institute; 2016.
CrossRef - Colgan, R., Keating, K., Dougouih, M. (2007). Survey of symptom burden in women with uncomplicated urinary tract infections. Clinical drug investigation 24(1), pp. 55–60.
CrossRef - Feng, Q.L., Wu, J., Chen, G.O., Cui, F.Z., Kim, T.N., Kim, J.O. (2000). A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mater. Res. 52:662–668.
CrossRef - Franke, S., Grass, G., Rensing, C. and Nies, D.H. (2001). The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology, 147(4):965-72.
CrossRef - Franke, S., Grass, G., Rensing, C. and Nies, D.H. (2003). Molecular analysis of the copper- transporting efflux system CusCFBA of Escherichia coli. J Bacteriol 185: 3804–3812.
CrossRef - Grass, G. and Rensing, C. (2001). Genes involved in copper homeostasis in Escherichia coli. J Bacteriol.;183(6):2145-7.
CrossRef - Grass,G.,Rensing,C.,and Solioz,M.(2011).Metallic copper as an antimicrobial surface. Appl. Environ. Microbiol. 77, 1541–1547.
CrossRef - Gupta, A., Phung, L.T., Taylor, D.E., Silver, S.(2001). Diversity of silver resistance genes in IncH incompatability group plasmids. Microbiology 147:3393–3402.
CrossRef - Johnson, T.J., Wannemeuhler, Y.M., Scaccianoce, J.A., Johnson, S.J., Nolan, L.K. (2006). Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob Agents Chemother 50:3929–3933.
CrossRef - Kim, J.S., Kuk, E., Yu, K.N., Park, S.J., Lee, H.J., Kim, S.H., Park, Y.K., Hwang, C.-Y., Kim, Y.-K., Lee, Y.-S., Jeong, D.H. and Cho, M.-H. (2007) ‘Antimicrobial effects of silver nanoparticles’, Nanomedicine: Nanotechnology, Biology and Medicine, 3(1), pp. 95–101.
CrossRef - Kittleson, J., Loftin, I., Hausrath, A., Engelhardt, K., Rensing, C. and McEvoy, M. (2006). Periplasmic metal-resistance protein CusF exhibits high affinity and specificity for both CuI and AgI’, Biochemistry., 45(37), pp. 11096–102.
CrossRef - Koowatananukul, C., Chansong, N. and Chuanchuen, R. (2010) ‘The contribution of active Efflux in reduced susceptibilities to copper Sulfate and zinc chloride in Escherichia coli isolates from swine’, Vet. Med, 40(3), pp. 317–321.
CrossRef - Loftin, I.R., Franke, S., Blackburn, N.J. and McEvoy, M.M. (2007). Unusual cu(I)/ag(I) coordination of Escherichia coli CusF as revealed by atomic resolution crystallography and x-ray absorption spectroscopy. Protein science 16(10): 2287–2293.
CrossRef - Lüthje, P., Brauner, A. (2014). Virulence factors of uropathogenic E. Coli and their interaction with the host. Advances in microbial physiology 65:337–72.
CrossRef - Macomber, L., Rensing, C. and Imlay, J.A. (2007) ‘Intracellular copper does not Catalyze the formation of Oxidative DNA damage in Escherichia coli’, Journal of Bacteriology, 189(5), pp. 1616–1626.
CrossRef - Nies, D.H. (2003). Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27: 313–339.
CrossRef - Pontel, L.B. and Soncini, F.C. (2009) ‘Alternative periplasmic copper-resistance mechanisms in gram negative bacteria’, Molecular Microbiology, 73(2), pp. 212–225.
CrossRef - Prabhu, S. and Poulose, E.K. (2012). Silver nanoparticles: Mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. International Nano Letters, 2(1), p. 32.
CrossRef - Rensing, C. and Grass, G. (2003). Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev 27: 197–213.
CrossRef - Reyes-Jara, A., Cordero, N., Aguirre, J., Troncoso, M. and Figueroa, G. (2016). Antibacterial effect of copper on microorganisms isolated from bovine Mastitis. Frontiers in Microbiology 7:626.
CrossRef - Roane, T.M. and Kellogg, S.T. (1995) .Characterization of bacterial communities in heavy metal contaminated soils. Can. J. Microbiol, 42, 593-603.
CrossRef - Sabry, S.A., Ghozlan, H.A. and Abou-zeid, D.M. (1997). Metal tolerance and antibiotic resistance patterns of a bacterial population isolated from sea water. J. Appl Microbiol, 82, 245-252.
CrossRef - Warnes,S.L.,Green,S.M.,Michels,H.T.,andKeevil,C.W.(2010).Biocidal efficacy of copper alloys against pathogenic enterococci involves degradation of genomic and plasmid DNAs. Appl. Environ. Microbiol. 76, 5390–5401.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





