How to Cite | Publication History | PlumX Article Matrix
Cloning and Characterization of Lipase Gene from A Local Isolate of Pseudoxanthomonas Sp.
Yogi Yopa Kristia1, Syifa F Syihab1 and Akhmaloka1,2
1Biochemistry Research Group, Faculty of Mathematics and Natural Sciences. Institut Teknologi Bandung, Jl. Ganesha no. 10 Bandung 40132, Indonesia.
2Departement of Chemistry, Faculty of Sciences and Computer. Universitas Pertamina, Jakarta, Indonesia.
Corresponding Author E-mail: loka@chem.itb.ac.id
DOI : http://dx.doi.org/10.13005/bbra/2471
ABSTRACT: Lipase gene from Pseudoxanthomonas sp. was cloned through in vitro amplification from total chromosomal DNA. The gene was sequenced and characterized, coding for 312 amino acid residues. Homological analysis showed that the gene has 98% similarity to lipolytic gene from Uncultured Pseudomonas sp (GenBank No. AKA58891.1). Further analysis appeared that the sequences showed similar unique motifs of lipase sub-family I.1, such as pentapeptide (GHSHG) motif, tetrapeptide (GMLG) motif, and catalytic triad. In additional, 3D structure analysis based on crystal structure of Pseudomonas aeruginose (PDB ID 1ex9) showed that both structure of lipases are similar except on the conformation of catalytic residue of His277 showing to shift more far away compared to that the control.
KEYWORDS: Cloning; Sequencing; Characterization P. seudoxanthomonas sp.; Thermostable lipase;
Download this article as:| Copy the following to cite this article: Kristia Y. Y, Syihab S. F, Akhmaloka A. Cloning and Characterization of Lipase Gene from A Local Isolate of Pseudoxanthomonas Sp. Biosci Biotech Res Asia 2017;14(2). |
| Copy the following to cite this URL: Kristia Y. Y, Syihab S. F, Akhmaloka A. Cloning and Characterization of Lipase Gene from A Local Isolate of Pseudoxanthomonas Sp. Biosci Biotech Res Asia 2017;14(2). Available from: https://www.biotech-asia.org/?p=25808 |
Introduction
Lipase is an enzyme that catalyzes hydrolysis of triacylglycerol into fatty acids and glycerol on the interface between water and organic solvent (Lotti et al., 2007). Lipases also show many catalytic activities besides hydrolisation such as esterification, trans-esterification, inter-esterification, acidolysis, aminolysis, alcoholysis and racemic resolution (Houde et al., 2004; Salihu and Alam, 2014, Sharma et al., 2001; and Brilliantoro et al., 2015). Since the enzymes show wide range activities, lipases are widely used in various fields of industries, such as food, detergents, cosmetic, biomedicine, biopolymers, biosurfactan, biodesel, and pharmaceutical industries (Jaeger and Eggert, 2002; Gupta et al., 2004; Houde et al., 2004; Salihu and Alam, 2014). Most of lipases used in industries are isolated from microorganism, since lipases from bacteria shown and an activity in wide range of temperature and pH (Akhmaloka et al., 2006).
Thermostability is desirable property for commercial lipase since enzymatic reaction at a high temperature could increase conversion process and the solubility of the substrats, reducing contamination and lowering the viscosity of the medium (Leow et al., 2004). Therefore, exploration of lipase from other sources have still extensively been carried out (Akhmaloka et al., 2006).
Some of thermophilic microorganisms have been isolated from compost (Madayanti et al., 2008; Nurhasanah et al., 2015; Syihab et al., 2015), hot spring and other sources (Akhmaloka et al., 2006; Syihab et al., 2015). Some of the isolated showed lipolytic activity (Widhiastuty et al., 2009; Febriani et al., 2013; Nurhasanah et al., 2017).
In previous paper (Syihab et al., 2015), we reported five isolates of lipolytic thermophiles with alcohol tolerance. In this paper cloning and characterization of one of the lipase will be described.
Materials and Methods
Chemicals
Common chemicals with pro analysis grade were purchased from Merck (Germany) and Sigma-Aldrich (USA). Bacterial growth nutrients, such as yeast extract, tryptophan were obtained from Bio Basic (Canada). Biochemical reagents such as dNTPs, PCR buffer, Taq DNA Polymerase were purchased from Fermentas (USA) and Kapa Biosystems (USA). Oligonucleotides (primers) were obtained from Macrogen (South Korea) and Integrated DNA Technologies (Singapore). Purification of PCR product used GeneJET Gel Extraction Kit (Thermo Scientific). The cloning was performed by using pJET1.2/blunt vector and T4 DNA ligase purchased from Promega (USA). Restriction enzymes were purchased from Thermo Scientific (USA).
Strain and Medium
Thermostable bacteria were obtained from our culture collections in the Laboratory of Biochemistry, Faculty of Mathematics and Natural Sciences, Institut Teknologi Bandung. The culture of Pseudoxanthomonas taiwanensis (AL17) was incubated in shaker incubator at 55oC with aeration rate at 150 rpm. Escherichia coli TOP 10 was used as host for gene cloning. The isolate was cultivated using modified media of Luria Berthani composed of 0.1% CaCl2.H2O, 0.5% yeast extract, 0,5% lab lemco/meat extract and 0,1% NaCl.
Isolation of Chromosomal DNA
Chromosomal DNA was isolated by using modified method of Zhou et al. (1996). The collected DNA pellet was separated from the supernatant and dried, subsequently followed by resuspension with 50µL ddH2O and stored at 4oC. The DNA was used for PCR.
PCR and Sub-Cloning of Lipase Gene
Cloning of lipase gene in each bacterial isolate was started by in vitro amplification of the gene by PCR technique using a pair of specific primers xFLipS2 (5’-ATGAACAAGAACAAAACCTTGCTCGCC 3’) and xRLipS2 (5’ TCAGAGCCCCGCGTTCTTCAA-3’) (Asy’ari et al., 2014). A typical PCR mixture (50 μL in volume) was prepared by mixing 5 μL of PCR buffer 10´, 2.5 mM MgCl2, 250 μM of deoxynucleosid-e triphosphate, 0.25 μM of each primers, and 1.25 U of Taq DNA polymerase. PCR was conducted by the following protocol: an initial denaturation was set at 98oC for 4 min followed by 35 cycles of denaturation (@30 s at 98oC), an annealing was programmed at 55oC for 30 s, while an extension and final extension were carried out at the same temperature at 72oC for 1 min and 5 min, respectively. The product of PCR was verified by electrophoresis technique using submerged horizontal electrophoresis gel for 45 min at 70 volts. The purification of PCR product was carried out by GeneJET Gel Extraction Kit. Finally, purified DNA was re-suspended to 50 μL buffers (10 mM Tris-HCl, pH 8.5). The purified DNA solution was stored at -20oC.
Construction of recombinant plasmid was carried out by ligating the PCR products with pJET1.2/blunt plasmid. The competent cell of E. coli Top 10 was prepared following the standar method. Transformation of E. coli was conducted by heat shock method. 100 mL of the transformed cells were spreaded on LB agar (LBA) media containing 100 mg/mL of ampicillin, 15 mg/mL of tetracycline and incubated at 37oC for overnight. Plasmid isolation from the transformed cell was carried out by the alkaline lysis method. Finally to verify the recombinant plasmid containing DNA insert, the plasmid was digested with Bgl II and visualized by agarose gel electrophoresis.
Sequencing of Lipase Gene
Lipase genes were sequenced based on Dideoxy- Sanger dye terminator method at Firstbase, Malaysia. The sequences were validated by analyzing the electrophoregram data using Sequence scanner 2 (Applied Biosystems). In order to combine the partial gene sequences into a full gene, we used DNA Baser Sequence Assembler v3 program (Heracle BioSoft).
Sequence Analysis
Deduced amino acid sequences of lipase were performed by in silico translation using Bioedit software and online server of ExPASy-Translate tool available at http:// web.expasy.org/translate/. Homologycal analysis were carried out using NCBI-Blastp analysis program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). A hundred of high homologous sequences were used to generate phylogenetic profile using MEGA 6 software based on the Neighbor-Joining clustering method. Amino acid composition and alignment analysis were performed using “Amino Acid Composition” and “ClustalW Multiple Alignment” programs which are integrated in the Bioedit software.
The analysis of lipase gene sequences was carried out by evaluating contents of GC, GCAT,and GC-AT of codon usage. The last two analyses were conducted with Bioedit-software,while the third analysis was calculated with an online program available at http:// www.bioinformatics.org/sms2/codon_usage.html. The analysis of codon usage was performed on the 1st, 2nd and 3rd base position. All analyzes above were not only performed on the isolate lipase sequences but also to other lipase genes from the other microorganism.
Result and Discussion
Cloning and Sequencing of Lipase Genes
Lipase genes have been successfully amplified from Pseudoxanthomonas sp (Figure 1) local strains by in vitro amplification. The gene, namely LipAL17 codes for 312 amino acids with length of 936 bp. The lipase gene was cloned into E. coli Top 10 and deposited to the GenBank database with accession number of ID 1918703, respectively.
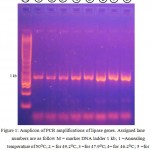 |
Figure 1: Amplicon of PCR amplifications of lipase genes. Assigned lane numbers are as follow M = marker DNA ladder 1 kb; 1 =Annealing temperature of 50°C; 2 = for 49.2°C, 3 =for 47.9°C; 4= for 46.2°C; 5 =for 43.9°C; 6 =for 41.9°C; 7= for 40.6°C and 8= for 40°C.
|
Homological of the Lipase
Homological analysis of deduced amino acid sequences showed that the gene appeared highly similarities to several lipases (Table 1), such as lipases from Uncultured Pseudomonas sp AKA 588891.1 and AKA 58893.1 and the Pseudomonas stutzeri AID 66451.1 and WPO 45159003.1 with percent identity of 98%.
Table 1: Homological result of Lip AL17 to 10 highest homolog. All of ten best homolog are lipases.
| No. | Description | Total score | Ident | Accession |
| 1. | lipase [uncultured Pseudomonas sp.] | 624 | 98% | AKA58891.1 |
| 2. | lipase [Pseudomonas stutzeri] | 624 | 98% | AID66451.1 |
| 3. | lactonizing lipase [Pseudomonas stutzeri] | 622 | 98% | WP_045159003.1 |
| 4. | lipase [uncultured Pseudomonas sp.] | 622 | 98% | AKA58893.1 |
| 5. | lactonizing lipase [Pseudomonas stutzeri] | 620 | 98% | WP_011913157.1 |
| 6. | lactonizing lipase [Pseudomonas stutzeri] | 619 | 97% | WP_019406249.1 |
| 7. | lactonizing lipase [Pseudomonas stutzeri] | 618 | 97% | WP_048329231.1 |
| 8. | lipase [Pseudomonas stutzeri] | 617 | 97% | WP_063543112.1 |
| 9. | lipase [Pseudomonas stutzeri] | 616 | 97% | WP_053528194.1 |
| 10. | LipA [Pseudomonas mendocina] | 613 | 96% | AAM14701.1 |
From 100 best homology sequences, the gene showed similar sequence with other lipases containing same conserved regions, such as GGGX, GXSXG (pentapeptide), oxyanion and catalytic triad (Ser, Asp, and His) (Figure 2). The presence of these conserved regions in the gene suggesting that the lipase is belonged to a member of family I.1. Lipase from sub-family I.1 have molecular masses in the range of 30-32 kDa and display a higher sequence similarity to the Pseudomonas aeruginosa (Jaeger and Eggert, 2002). The subfamilies I.1 also share important structural features. A part from the residues forming the catalytic triad, two aspartic residues involved in the Ca2+-binding site (Nardini et al., 2000). Two cysteine residues forming a disulphide bridge are conserved in a majority of the sequence (Nardini et al, 2000). Residues involved in the formation of both the Ca2+-binding site and the disulphide bridge are located in the vicinity of the catalytic His and Asp residues (Jaeger et al., 1999) and believed to be important role on the stabilization of the active center of the enzymes.
 |
Figure 2: Conserved region of lipase covers of GMLG (tetrapeptide), GHSHG (pentapeptide), oxyanion and catalytic triad in 9 highest lipase sequences. A description of the figure is as follows: A. position of signal peptide; B. tetrapeptide motif; C. pentapeptida motif; catalityc triad; Asp residue; Arg residue
|
For further characterization 3D structure of the lipase was constructed in silico based on 3D structure of the P. aeruginosa (POB ID I EX 9). The superimposed result showed that 3D structure of LipAL17 and I EX 9 are similar (Figure 3) except on the conformational of His277 in the active center of the enzyme that is believed to have an impact on the activity of the enzyme (Nurhasanah et al., 2015; Nurhasanah et al., 2017)
 |
Figure 3: Crystal structure comparison between Lip AL17 and I EX 9. A. Model Structure of Lip AL17; B. Crystal Structure of I EX 9; C. Superimposed between Lip AL17 and I EX 9, His277 Lip AL17 showed to shift more far away compared to that the control (I EX 9).
|
Acknowledgements
This research was supported by research grant from The Ministry of Research, Technology, and Higher Education of The Republic of Indonesia.
References
- Akhmaloka., Nurbaiti S., Hertadi R., Yohandini H., Aminin A. L., Helwati H and Madayanti F. Exploration of thermophilic microorganism from hot springs around Java. Journal of the Indonesian Chemical Society. 2006;1:1-9.
- Brilliantoro F., Zidny R., Widhiastuty M. P and Akhmaloka. Hydrolitic and Transesterification Activities of Thermostable Lipase ITB 1.1. Biosciences Biotechnology Research Asia. 2015;12(1):01-06.
- Febriani., Ihsanawati., Akhmaloka., Hertadi R and Warganegara F. Thermostable alkaline Lipase isolated from Thermus aquaticus of the Kamojang. International Journal of Integrative Biology. 2013;14(2):104–112.
- Gupta R., Beg Q. K and Lorenz P. Bacterial Alkaline Proteases: Molecular Approaches and Industrial Applications. Appl Microbiol Biotechnol. 2002;59:15-22.
CrossRef - Houde A., Kademi A and Leblane D. Lipases and Their Industrial Applications: An Overview. Applied Biochemistry and Biotechnology. 2004;118:155-170.
CrossRef - Jaeger K. E., Dijkstra B. W and Reetz M. T. Three-Dimensional Structures and Biotechnological Applications of Lipases. Annual Review of Microbiology. 1999;53:315-351.
CrossRef - Jaeger K. E and Eggert T. Microbial Lipases Form Versatile Tools for Biotechnology; Review. Trends Biotechnol. 1998;16:396-403.
CrossRef - Leow T. C., Rachman R. N. Z. R. A., Basri M., dan Salleh A. B. High Level Expression of Thermos table Lipase from Geobacillus sp. Strain T 1. Biotech. Biochem. 2004;68(1):96-103.
CrossRef - Lotti M and Alberghina L. Lipases: Molecular Structure dan Function. Polaina dan A. P. MacCabe (eds.), Industrial Enzymes. 2007;263–281.
CrossRef - Madayanti F., Viera B. V. E., Whidiastuty M. P and Akhmaloka. Characterization and Identification of Thermophilic Lipase Producing Bacteria from Thermogenic Compost. Journal of Pure and Applied Microbiology. 2008;2(2):325-332.
- Nardini M., Lang D. A., Liebeton K., Jaeger K and Dijkstra B. 2000.
- Protein Structure and Folding: Crystal Structure of Pseudomonas aeruginosa Lipase in the Open Conformation: the prototype for Famili I.1 of Bacterial Lipases. The Journal of Biological Chemistry. 275(40):31219-31225.
CrossRef - Nurhasanah., Nurbaiti S., Madayanti F., Akhmaloka. Diversity of Gene Encoding Thermostable Lipase from Compost Based on Metagenome Analysis. International Journal of Integrative Biology. 2015;16(1):07-12.
- Salihu A and Alam Md Z. Thermostable Lipases an Overview of Production Purification and Characterization. Biosciences Biotechnology Research Asia. 2014;11(3):1095-1107.
CrossRef - Sharma R., Chisti Y and Banerjee U. C. Production Purification, characterization, dan applications of lipases. Research review paper. Elsevier. Biotechnology Advance. 2001;627-662.
- Syihab S. F., Madayanti F and Akhmaloka Isolation, Characterization and Identification of Lipolytic Thermophiles with Methanol Tolerance from Domestic Compost. Journal of Pure and Applied Microbiology. 2015;9(2):385-390.
- Widhiastuty M. P., Febriani H.,Yohandini M. R., Madayanti F. M and Akhmaloka. Characterization and Identification of Thermostable Alkaline Lipase Producing Bacteria from Hot Spring around West Java. Journal of Pure and Applied Microbiology. 2009;3(1):27-40.
- Zhou J., Bruns M. A., dan Tiedje J. M. DNA Recovery from Soils of Diverse Composition. Applied and Environmental Microbiology. 1996;62(2):316-322.

This work is licensed under a Creative Commons Attribution 4.0 International License.





