How to Cite | Publication History | PlumX Article Matrix
Samira Dehnavi and Bahareh Rahimian Zarif
Department of Biology, Sanandaj Branch, Islamic Azad University, Sanandaj, Iran.
Corresponding Author E-mail: Bahareh_r_z@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2511
ABSTRACT: Urinary tract infections are among prevalent diseases which can happen in women and men in different age groups. Proteus mirabilis is considered as the cause of 10 percent of urinary tract infections and is the fifth common nosocomial urinary tract infection. A major problem in nosocomial infections is emergence of multidrug resistant species. It is assumed that rsbA in Proteus mirabilis is an engaged gene in this process. Consequently, in this study we have assessed the relation between the presence of this gene and antibiotic resistance. 40 urine sample which were diagnosed to be positive for Proteus mirabilis using differential test, were assessed for antibiotic resistance to beta lactam antibiotics using diffusion disk and were analyzed using PCR for rsbA gene. Results of antibiotic resistance and presence of rsbA gene were analyzed using Pearson correlation statistical test. Results showed 60%, 50%, 34%, 44%, and 20% antibiotic resistance for penicillin, cefotaxime, carbapenem, cefipenem, and ceftriaxone respectively. RsbA gene was present in 67% of samples. Pearson statistical analysis showed a statistically significant relationship between antibiotic resistance and rsbA gene.
KEYWORDS: antibiotic resistance; Proteus mirabilis; rsbA gene; urinary infection
Download this article as:| Copy the following to cite this article: Dehnavi S, Zarif B. R. RsbA Gene's Confirmation in P. Mirabilis With PCR Method in Patients With Urinary Tract Infection in Province of Qorveh. Biosci Biotech Res Asia 2017;14(2). |
| Copy the following to cite this URL: Dehnavi S, Zarif B. R. RsbA Gene's Confirmation in P. Mirabilis With PCR Method in Patients With Urinary Tract Infection in Province of Qorveh. Biosci Biotech Res Asia 2017;14(2). Available from: https://www.biotech-asia.org/?p=25305 |
Introduction
Urinary tract infections (UTI) are among prevalent diseases which can happen in women and men in different age groups. Approximately 30 percent of women experience UTI requiring treatment before 24 and about half women experience symptoms of UTI in their life. Urinary system is usually sterile and no bacteria can be found in it but colonized bacteria in perineal region which are mostly present bacteria in rectum may enter bladder via urinary tract.1
Usually, host defense mechanisms prevent form bacteria growth and duplication and occurrence of UTI. When there are too many bacteria or their virulence is worsen or the defense of host in urinary tract is suppressed, UTI may occur. UTI is defined as an inflammatory response of urothelium to bacterial invasion which is associated with bacteriuria and pyuria.2
Most UTI cases are caused by bacteria which are part of intestinal, vaginal, or perineal skin flora or other Enterobacters such as Proteus or Klebsiella.
The most common gram positive bacteria causing UTI is Staphylococcus saprophyticus or Staphylococcus aureus. Uncommon bacteria such as Citrobacter, Serratia, Pseudomonas, and providencia are mostly detected in nosocomial infections.3
UTI are among recurrent bacterial infections in human and account for about 20 percent of diseases out of hospital. Approximately, 90 percent of UTIs are progressively ascending, that is, they enter upper parts of urinary tract via urinary bladder.1 Proteus mirabilis is considered the cause of 10 percent of UTIs and the fifth common cause of nosocomial urinary infections. This bacteria places on urinary catheters and by forming biofilms, results in obstruction in urinary tracts. Many hospitalized patients are infected by catheters and it is considered a major concern by emergence of MDR strains.4
An outstanding characteristic of Proteus mirabilis is its ability to crawl on the agar and form very orderly edged colonies with bulls-eye appearance. Crawling of this bacteria is the result of periodic differentiation of vegetating cells to crawling cells.5
Proteus infection stability depends on the ability of the bacteria to form urinary stone and placing on the catheters. 6 Because, this way, it can be kept safe form immune system and antibiotics. In fact, stone formation around the microorganism prevents form effect of immune system and medications on the microorganism and these strains are resistant to cephalosporin.7-8
A gene condign virulence factor is rsbA whose product is the controller of swarming motility in Proteus which acts as a sensor of environmental conditions.9 RsbA protein leads to formation of biofilms and extracellular polysaccharides in this bacteria.10
In the conducted studies on beta lactam resistant species, it was found that Proteus strains with more swarming motility show higher resistance rate to beta lactam medications.11
In this study, we assess presence of this gene in samples of Proteus mirabilis UTI and its relationship with antibiotic resistance.
Materials and Methods
50 urine samples with verified infection with Proteus mirabilis, were used. 0.1 ml of urine sample and inoculated on Macconkey culture medium and incubated in 37 degrees centigrade for 24 hours. Since this bacteria is not able to deferment lactose, it forms colorless colonies on Macconkey culture medium. Formed colonies were passaged for isolated culture. To diagnose this isolates and verify genus and species of the bacteria, standard biochemical tests were performed on the samples. First, gram staining, oxidase and catalase test were performed on pure isolates their identity was proved by culturing on simon citrate agar, urea agar, triple sugar iron agar culture media, motility test, indole test, gas forming test, vags prosecco test, citrate test, and urease test. Antibiotic resitance pattern were determined by kirby bauer disk infusion test. First, monoclonal isolates were prepared. After 18-24 hours, microbial isolates were dissolved in physiologic serum with sterile ansell until its turbidity is similar to half macfarland solution. Then meadow cultivation was performed on Mueller-Hinton agar using sterile swab with standard distribution and the disks were put in appropriate places and plates were put in 37 degrees centigrade incubator. Results of tests were read 18 hours later and the radius of hallow of lack of growth was analyzed.
Table 1: Sequences of Primers used for rsbA gene amplification
| Name of Primer | Sequence of primer |
| rsbAF | TTGAAGGACGCGTCAGACC |
| rsbAR | ACTCTGCTCCTGTGGGTA |
Results
Results of antibiotic resistance are showed in table 2. DNA was extracted from all samples and quality of extracted DNA were analyzed by OD280 assessment and sing agarose gel which showed proper quality of extracted samples. These samples were assessed for rsbA gene. PCR program for this test is shown in table 3 and PCR product is depicted in figure 1. Results of PCR for rsbA gene showed that 68% of samples had rsbA gene.
To evaluate the relationship between antibiotic resistance and presence of rsbA gene in Proteus mirabilis, Pearson coefficient of correlation statistical test was used. In 34 cases of a total of 50 samples, rsbA gene was amplified. 26 cases of rsbA positive samples showed more than 50 percent antibiotic resistance. Results of Pearson coefficient of correlation suggests statistically significant relationship between antibiotic resistance and expression of rsbA gene (P-value<0.05).
Table 2: antibiotic resistance in Proteus mirabilis cultures
| Name of antibiotic | Resistance percentage |
| Penicillin | 60% |
| Cefotaxime | 50% |
| Carabpenem | 34% |
| Cefipenem | 44% |
| Ceftriaxone | 20% |
 |
Figure 1: product of bacterial gene extraction on agarose gel
|
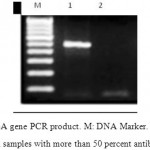 |
Figure 2: rsbA gene PCR product. M: DNA Marker. 1: rsbA gene PCR product in samples with more than 50 percent antibiotic resistance. 2: rsbA gene PCR product in samples with less than 50 percent antibiotic resistance
|
 |
Table 3: results of Pearson coefficient of correlation Click here to View table |
Discussion
In the current study, 50 urine samples with Proteus mirabilis infection were analyzed and checked for rsbA gene in positive samples. rsbA gene is one of the genes involved in swarming phenomena which is characteristic for Proteus mirabilis. In 2004, Jen Liaw Shwu et al. proved the presence of rsbA gene in Proteus mirabilis.12 They proved the presence of rsbA gene, its mutants and its role in swarming phenomena and expression of virulence factor in wild strain of Proteus mirabilis, P19. Their results were consistent with Belas et al. study in 1998.13
In evaluated samples in this study, 68 percent of them were positive for rsbA gene. In Ahmadi et al. study in 2014, they stated that rsbA gene was present in 70 percent of animal samples which was consistent with this study.14
Resistance to beta lactam antibiotics were included in this study. Result showed a relatively high rate of resistance.
One of the goals of this study was to discuss the relation between antibiotic resistance and the expression of rsbA gene. Resistance rate was defined as 0 or 1 in this study. Zero was devoted to samples with under 50 percent resistance and one was devoted to samples with above 50 percent resistance. Then the correlation of antibiotic resistance and presence of rsbA gene were analyzed using Pearson coefficient of correlation. Result showed a statistically significant relationship.
Production of beta lactamase is the reason of beta lactam antibiotic resistance but biofilm formation and swarming ability let the bacteria escape form the effect of bacteria. Shwu-je Liaw studies in 2001 and Ahmadi in 2014 verified the role of rsbA gene in antibiotic resistance.12-14 They showed that rsbA gene plays a double role in swarming regulation and biofilm formation. Stewart et al. stated in their study in 2002 that biofilm formation in bacteria is one of the most important causes in antibiotic resistance and for defending against bacteria with this capability, responsible products and genes should be targeted.15
In 2003, Xaveir et al. showed in their study that rsbA gene plays role in qourom sensing phenomena.16 This procedure, means using (self-inducing) signals, for communicating with other bacteria and consequently in organized expression of gene, namely qourom sensing, is the cause of antibiotic resistance. It has been shown that biofilms have high antibiotic resistance and are considered as one of the most important therapeutic problems and qourom sensing plays a crucial role in biofilm formation.
According to growing antibiotic resistance which is now a great concern for physicians, therapeutic methods which cut down on antibiotic consumptions are at the center of attentions. Based on the importance of qourom sensing phenomena in bacterial virulence, next generation of antibiotics should target qourom sensing. In Hoiby et al. study in 2010, it was stated that proper solution to encounter bacteria with the ability to form biofilm and used qourom sensing is using inhibitors of this phenomena or using substances which degrade biofilm, such as DNase and gelatinase.17
Also, we may suppose using these inhibitors with other medications with synergism effect and lower antibiotic dosage. In some cases, drug sensitivities does not let using antibiotic. Maybe, this can be attenuated with emergence of the next generation of these drugs.
Finally, it can be concluded that Proteus mirabilis bacteria is the cause of 10 percent of nosocomial infections due to expression of rsbA gene and biofilm formation ability, qourom sensing, and consequent antibiotic resistance. Effective medications can be designed for greater success rate.
References
- Lane D. R., Takhar S. S. Diagnosis and management of urinary tract infection and pyelonephritis. Emergency medicine clinics of North America. 2011;29(3):539-52.
CrossRef - Abraham S. N., Miao Y. The nature of immune responses to urinary tract infections. Nature Reviews Immunology. 2015;15(10):655-63.
CrossRef - Sheerin N. S. Urinary tract infection. Medicine. 2011;39(7):384-9.
CrossRef - Jones H. E., Park R. W. The influence of medium composition on the growth and swarming of Proteus. Microbiology. 1967;47(3):369-78.
- Nys S., Van Merode T., Bartelds A. I., Stobberingh E. E. Urinary tract infections in general practice patients: diagnostic tests versus bacteriological culture. Journal of Antimicrobial Chemotherapy. 2006;57(5):955-8.
CrossRef - Mobley H. L., Warren J. W. Urease-positive bacteriuria and obstruction of long-term urinary catheters. Journal of clinical microbiology. 1987;25(11):2216-7.
- Bichler K. H., Eipper E., Naber K., Braun V., Zimmermann R., Lahme S. Urinary infection stones. International journal of antimicrobial agents. 2002;19(6):488-98.
CrossRef - Rózalski A., Sidorczyk Z., Kotełko K. R. Potential virulence factors of Proteus bacilli. Microbiology and Molecular Biology Reviews. 1997;61(1):65-89.
- O’Hara C. M., Brenner F. W., Miller J. M. Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clinical microbiology reviews. 2000;13(4):534-46.
CrossRef - Manos J., Belas R. The genera Proteus Providencia and Morganella. In The prokaryotes Springer New York. 2006;245-269.
- O’Hara C. M., Brenner F. W., Steigerwalt A. G., Hill B. C., Holmes B., Grimont P. A., Hawkey P. M., Penner J. L., Miller J. M., Brenner D. J. Classification of Proteus vulgaris biogroup 3 with recognition of Proteus hauseri sp. nov., nom. rev. and unnamed Proteus genomospecies 4, 5 and 6. International journal of systematic and evolutionary microbiology. 2000;50(5):1869-75.
CrossRef - Liaw S. J., Lai H. C., Wang W. B. Modulation of swarming and virulence by fatty acids through the RsbA Protein in Proteus mirabilis. Infect Immun. 2004;72:6836–45.
CrossRef - Belas R., Erskine D., Flaherty D. Proteus mirabilis mutants defective in swarmer cell differentiation and multicellular behavior. J Bacteriol. 1991;173:6279–88.
CrossRef - Ahmadi S., Norozi J., Sepahi A. evaluation the relation between rsb A gen and meristic acid in virulence of proteus mirabilis extracted from urin. Research in medicine. 38(3);162-166.
- Stewart P. S. Mechanisms of antibiotic resistance in bacterial bio films. International Journal of Medical Microbiology. 2002;292(2):107-13.
CrossRef - Xavier K. B., Bassler B. L. Lux S quorum sensing more than just a numbers game. Curr Opin Microbiol. 2003;6:191–97.
CrossRef - Høiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. Antibiotic resistance of bacterial bio films. International journal of antimicrobial agents. 2010;35(4):322-32.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





