How to Cite | Publication History | PlumX Article Matrix
Mushtaq T. Sh. Al-Rubaye1, Mastafa H. J. Al-Musawi2, Javad Fakhari1 and Maryam Hosseini1
1Department of Microbiology, Faculty of Biological Sciences, Shahid Beheshti University, Tehran, Iran.
2College of Pharmacy, University of Al-Mustansriyah, Baghdad, Iraq.
Corresponding Author E-mail: maryamhosseini83@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2476
ABSTRACT: A total of 218 halophilic bacterial isolates were obtained from Lake Razazah, west of Karbala, Iraq. Optimum pH and temperature were 7.5 and 37 °C, respectively. According to optimal growth at different salt concentration, the slightly halophilic bacteria were the most abundant isolates with the frequency of 68%. The isolated bacteria were screened for the production of extracellular amylase, alkaline amylase, protease, alkaline protease, lipase, alkaline lipase, pectinase and cellulase. The production of pectinase (55.8%), amylase (52.6%) and lipase (50.0%) were observed in almost half of the halophilic bacteria. Alkaline amylase and alkaline lipase production were reported in less than one third (30%) of isolates. Phylogenetic analysis16S rRNA sequences indicated that all isolates were members of eight genera of the domain Bacteria, including Bacillus, Halobacillus, Virgibacillus, Oceanobacillus, Staphylococcus, Pseudomonas, Idiomarina and Halomonas. The predominant commercial enzymes producers in current study were Halobacillus sp. K51 and Halomonas sp. K46 with the ability to produce 7 out of 8 exoenzymes. The presented data shows that despite drought, dehydration, increased concentrations of salt and contaminants, Lake Razazah represents an untapped source of halophilic bacteria biodiversity.
KEYWORDS: Biodiversity; Halophiles; Hydrolases; Isolation
Download this article as:| Copy the following to cite this article: Al-Rubaye M. T. S, Al-Musawi M. H. J, Fakhari J, Hosseini M. Screening and Characterization of Halophilic Bacteria With Industrial Enzymes from Salt Lake Razazah, Karbala, Iraq. Biosci Biotech Res Asia 2017;14(2). |
| Copy the following to cite this URL: Al-Rubaye M. T. S, Al-Musawi M. H. J, Fakhari J, Hosseini M. Screening and Characterization of Halophilic Bacteria With Industrial Enzymes from Salt Lake Razazah, Karbala, Iraq. Biosci Biotech Res Asia 2017;14(2). Available from: https://www.biotech-asia.org/?p=26078 |
Introduction
One of the widely distributed aquatic ecosystems is saline environments.1 Despite the high concentration of salt, a great diversity of life forms can be observed in this environment and they found in all the three domains of life, Bacteria, Archaea, and Eukarya.2 Halophiles include prokaryotic and eukaryotic salt-loving microorganisms of saline environments which are able to balance the high osmotic pressure.3 To adapt to high level saline, they have developed a number of biochemical strategies to maintain cell structure and function.4-6 The diversity amongst hamophiles regards to the concentration of salts, temperatures, pH conditions, and redox conditions that the organisms are adapted. Thermophilic, psychrophilic, mesophilic, alkaliphilic, aerobic and anaerobic halophilic bacteria have been described.,4-7 Halophiles play an important role in the carbon and phosphorus cycles in saline environments.8 Some of them are able to degrade organic compounds, such as crude oil which makes them potential candidates in bioremediation studies.9-10
Halophiles are diverse with regard to the concentration of salt which they are adapted. They are able to produce hydrolytic enzymes in hypersaline environment which possess a potential importance in industrial area.11 Moderately halophilic bacteria represent a group of microorganisms that is widely distributed in salinity zones. They show optimum growth at 5-15% NaCl.12-13 The bacteria grow at salt concentrations above 20% (w/v) to saturation named extremely halophilic.3 Slightly halophilic microorganisms are capable of growing optimally in media containing 2-5% NaCl.14
A variety of industrial applications of microbial hydrolase including textile, food, dairy, pharmaceutical, detergents, cosmetic industries and synthesis of polymeric materials were described in previous studies.15-18 Microbiological sources of extracellular enzymes are used for a wide range of industrial and biotechnological aims.12,19 Many recent studies have focused on extracellular hydrolytic enzymes of the halophilic bacteria in order to use such enzymes in biotechnological processes.20
Iraq features various ecosystems including extreme environments such as high salinity soil and water in which microbial diversity has been poorly studied.21-24 Lake Razazah, also known as Bahr Al-Milh (literally Sea of Salt) is located 10 kilometers west of Karbala province, Iraq (32°45′N 43°38′E). The covered area ranges from 1050 to 1700 km2depends on water evaporation through a year.24 The water level changes with the seasons but totally, the lake is rather shallow with maximum depth of 17 m. The pH values ranges from 7.1 to 8.5.25 Due to rapid evaporation of water, especially in summer, the water in this lake is quite saline and in different parts the salinity varies between 7.8 to 11%2.6-27 Lake Razazah fed by the Euphrates River via Sin-Al-Thibban canal.28 In current study, we identified the microbial diversity of Razazah Lake for the first time in this country. Screening of slightly, moderately and extremely halophilic bacteria were performed using soil, sludge and water samples of the lake. The prevalence of microorganisms producing extracellular hydrolytic enzymes was also described.
Material and Methods
Sample Preparation
Saline soil and water samples were collected from fifteen different zones, at the surface and various depths of the south and southeast of Razazah Lake (between longitude 42º 00′ – 43º 45′ E and latitude 32º 00′ – 33º 15′ N) on 26thFebruary 2014 (Figure 1). The samples were put into labeled plastic bags or sterile glass jars and transported to the laboratory for further analysis. Even though it was winter, the temperature was between 22 to 25 °C. All samples were cultured in nutrient broth with saline concentration of 2.5%, 5.0%, 7.5%, 10.0%, 12.5%, 15.0%, 17.5%, 20.0% and 22.5%. Sea salt consisted of 175 g NaCl, 20 g MgCl2.6H2O, 5 g K2SO4, 0.1 g CaCl2.2H2O and 2 mL filtered saline lake water (as source of trace elements) in 1 L distilled water. The culture media were adjusted to pH of 7.0, 7.5, 8.0 and 8.5 using NaOH. After autoclaving, all media were incubated at 37°C in orbital shaker, at 120 rpm for 7 days. Cell morphology was examined by using light microscopy (model BH 2; Olympus). Gram staining and KOH lysis tests were carried out according to previous studies.29-30
![Figure 1: Map of sampling sites at the south and southeast of Lake Razazah, Karbala, Iraq. The map was derived from Satellite World Map in 2014 [31]. Red arrows show the sampling sites.](https://www.biotech-asia.org/wp-content/uploads/2017/06/Vol_14_no2_Scr_Mus_fig1-150x150.jpg) |
Figure 1: Map of sampling sites at the south and southeast of Lake Razazah, Karbala, Iraq. The map was derived from Satellite World Map in 2014 [31]. Red arrows show the sampling sites.
|
Hydrolase Activities
Screening of bacteria based on extracellular hydrolase activities including amylase, alkaline amylase, protease, alkaline protease, lipase, alkaline lipase, pectinase and cellulase (CMCase) were performed using agar plates. In order to preparing alkaline media, pH was adjusted using NaOH.
Extracellular Protease Activity
A 10% skim milk-agar medium supplemented with a range of sea water concentrations was prepared by dissolving 10 g of skim milk powder in 100 mL of sterile nutrient agar. Ability to create clear zone around the colonies after incubation at 37°C for 7 days was considered as positive proteolytic activity.32
Extracellular Lipase Activity
Lipase activity was detected using sterilized basal nutrient agar media supplemented a range of sea water concentrations and containing 2.5% olive oil (w/v) at 60°C. Then, 0.001% rhodamine B was added to homogenized mixture at 50°C and poured. Observation of shinny colonies under UV irradiation after incubation of bacteria at 37°C for 7 days indicated lipolytic activity.33
Extracellular Amylase Activity
Extracellular amylase activity was assayed using 1% starch agar medium (Merck) using various concentration of sea water. To detect the hydrolysis of starch, I2–0.6%KI solution or Lugol’s iodine was used. After incubation at 37°C for 7 days, presence of dark blue, purple, or black color represented amylolytic activity.33
Extracellular CMCase Activity
Nutrient medium with 1% carboxymethyl cellulose (CMC) containing a range of artificial sea water were incubated at 37°C for 7 days. Clear zone around the colony after flooding in 0.1% congo red solution for 15 minutes showed CMCase activity.34
Extracellular Pectinase Activity
Sterile nutrient medium containing 1% pectin and varying concentration of sea water was used. Cultured media were incubated at 37°C for 7 days and then flooded in 0.3% I2–0.6%KI solution. Presence of clear zones around the growth was considered as pectinolytic activity.35
Molecular Analysis
Genomic DNA was extracted by Bioron sample preparation kit (Korea). The 16S rRNA gene was amplified by PCR using the universal prokaryotic primers including 8F (5´-AGAGTTTGATCCTGGCTCAG-3´) and 1492R (5´-GGTTACCTTGTTACGACTT-3´). PCR mixtures of 50 μL contained 17 μL dH2O, 1 μL primers, 1 μL dNTP, 0.75 μLMgCl2 (50mM), 25 μL PCR amplification buffer (1X), 2 U Taq DNA polymerase, 5 μL template DNA. The amplification was done by initial denaturation 95°C for 5 min, 30 cycles at 95° C 1 min, 58°C 30 s, 72°C 1 min with a final extension of 72° C for 10 min. Techne TC-3000X Thermal cycler (UK) was used for amplification. PCR products were electrophoresed in 1% (w/v) agarose gels and the single ~ 1400 bp of DNA fragments were purified using GeneAll Gel Extraction Kit (Korea). PCR products of the nearly complete 16S rRNA gene were sequenced in both directions using an automated sequencer by Macrogen Biotechnology Company (Korea).
The 16S rRNA gene sequence was aligned using BLAST and Ribosomal Database Project (RDP) with the published sequences of closely related bacteria in the GenBank database of the National Center for Biotechnology Information. Phylogenetic trees were constructed with the aid of the MEGA 7 software package36 using the neighbour-joining,37 maximumlikelihood38 and maximum-parsimony39 algorithms. The confidence values for the branches of phylogenetic trees were determined using bootstrap analyses based on 1000 replications.40 The 16S rRNA gene sequences of the strains have been deposited in the GenBank database.
Results and Discussion
The salinity of the Euphrates River has increased gradually to more than double over the last 30 years.27 Screening isolates from saline Lake Razazah under aerobic conditions led to isolation of 218 isolates from 15 samples. Optimum pH and temperature were 7.5 and 37°C, respectively. The slightly halophilic bacteria were the dominant isolates with the frequency of 68% followed by 32% moderately halophilic bacteria. They consist of 31 gram negative rods, 26 gram negative cocci, 95 gram positive rods and 66 gram positive cocci. No extremely halophilic bacteria were found. About two third (67.4%) of the isolates were halotolerant and only eighteen isolates grew at NaCl concentrations greater than 15.0%. Additionally, 15 protozoa, 3 algae, 5 fungi and yeast were observed (the data not shown).
Extracellular Hydrolase Production
The priority for further molecular analysis was the number of extracellular enzymes that has produced by the halophilic bacteria. The production of eight hydrolytic exoenzymes by twenty-one isolates was illustrated in table1. Halobacillus isolates were the most valuable species in terms of industrial enzyme production in the present study. Halomonas, Staphylococcus and Pseudomonas isolates showed similar strength in the production of the very exoenzymes. Halobacillus sp. K51 and Halomonas sp. K46 were able to produce 87.5% of abovementioned industrial enzymes. All Halobacillus and Bacillus isolates showed the ability of extracellular pectinase production. Halobacillus, Bacillus, Halomonas and Staphylococcus genera had isolates which were able to produce both extracellular amylase and alkaline amylase. The ability to produce extracellular protease and alkaline protease both were observed in isolates of Halobacillus, Virgibacillus, Bacillus, Halomonas and Staphylococcus. In addition, Halobacillus, Halomonas and Staphylococcus isolates were found to be potent lipase and alkaline lipase producers.
Table 1: Hydrolytic activities of the representatives of eight genera including Bacillus, Halobacillus, Virgibacillus, Oceanobacillus, Staphylococcus, Pseudomonas, Idiomarina and Halomonas isolated from Lake Razazah, Karbala, Iraq.
| Isolates | GenBank Accession Number | Amylase | Alkaline Amylase | Protease | Alkaline Protease | Lipase | Alkaline Lipase | Pectinase | Cellulase | Enzyme production (%) |
| Halobacillus sp. K13 | KT353097 | – | – | – | + | + | + | + | – | 50.0 |
| Halobacillus sp. K15 | KT991680 | + | – | – | + | + | – | + | – | 37.5 |
| Halobacillus sp. K16 | KT597929 | + | + | – | + | – | + | + | + | 75.0 |
| Halobacillus sp. K17 | KT353098 | – | + | + | + | + | – | + | – | 62.5 |
| Halobacillus sp. K51 | KT223788 | + | + | + | – | + | + | + | + | 87.5 |
| Virgibacillus sp. K11 | KR347118 | – | – | + | + | + | – | + | + | 62.5 |
| Virgibacillus sp. K19 | KT597930 | + | – | – | – | + | – | – | – | 25.0 |
| Oceanobacillus sp. K30 | KT281118 | – | + | – | + | – | – | – | – | 25.0 |
| Oceanobacillus sp. K31 | KT281119 | – | + | + | – | – | – | + | + | 50.0 |
| Bacillus sp.K3 | KT597927 | + | – | + | – | – | – | + | – | 37.5 |
| Bacillus sp.K14 | KT597928 | + | + | – | – | – | – | + | – | 37.5 |
| Bacillus sp.K21 | KT200230 | + | – | + | + | – | – | + | – | 62.5 |
| Bacillus sp.K32 | KT223787 | + | + | + | + | + | – | + | – | 75.0 |
| Bacillus sp.K73 | KT223786 | – | + | + | – | – | + | + | – | 50.0 |
| Halomonas sp. K29 | KT353099 | + | – | + | – | – | + | – | + | 50.0 |
| Halomonas sp. K40 | KT597931 | + | + | – | – | + | – | – | – | 37.5 |
| Halomonas sp. K42 | KR909223 | – | + | + | + | + | + | – | + | 75.0 |
| Halomonas sp. K46 | KT597932 | + | – | + | + | + | + | + | + | 87.5 |
| Staphylococcus sp. K33 | KT353100 | + | + | + | + | – | + | – | + | 62.5 |
| Idiomarina sp. K39 | KT200229 | + | – | – | + | – | – | – | + | 37.5 |
| Pseudomonas sp. K80 | KT597933 | + | – | + | – | + | – | + | + | 62.5 |
In this study, among all isolated halophilic bacteria of Lake Razazah (N=218), production of pectinase (55.8%), amylase (52.6%) and lipase (50.0%) were observed in half or more than half of the halophilic bacteria. On the other side, only less than one third of isolates were able to produce alkaline amylase and alkaline lipase. Near to half of them showed the ability of protease and alkaline protease production (Figure 2). In contrast with the study of Rohban et al., no lipase and amylase activity was reported for Oceanobacillus isolates.41 Kumar, S. et al. introduced Bacillus isolates as halophilic microorganisms with the maximum ability of lipase and protease productionfrom soil and water samples in India.42 Similar to our results, they found Oceanobacillus strains showed extracellular protease activity. Halomonas isolates from the Tunisian Solar Saltern also showed hydrolase producing ability.43 The environmental isolates of Salinivibrio, Halomonas, Bacillus and Salibacillus genera were predominant commercial enzymes producers in hypersaline environments of Spain.12,44
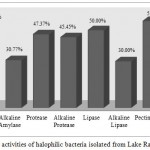 |
Figure 2: Hydrolytic activities of halophilic bacteria isolated from Lake Razazah, Karbala, Iraq
|
Amylase and alkaline amylase derived from the genus Bacillus is the main source of these enzymes with commercial applications.45 Similarly, in the present study, alkaline amylase activity was mainly observed in Bacillus, Halobacillus and Oceanobacillus isolates. In the study of the hypersaline environments in South Spain, only two isolates of Halomonas and Nesterenkonia genera were detected amylase positive.44 However, extracellular amylase able to be active in pH more than 8.0 was purified from other microorganisms.
Among our isolates, Halobacillus, Virgibacillus, Oceanobacillus, Staphylococcus, Idiomarina and Halomonas showed the protease activity in pH 8.5. An extracellular alkaline protease of an extreme halotolerant Bacillus was introduced from West Sea in Inchon, Korea.46 Same protease activity was observed in a haloalkaliphilic Bacillus sp. isolated from a seawater sample in Gujarat, India.47 An extracellular protease was produced under conditions of high salinity and pH 8.5 by a moderately halophilic Salinivibrio sp. isolated from Bakhtegan Lake, South of Iran.48
The Staphylococcus sp. K33 showed alkaline lipase activity similar to a newly soil-isolated Staphylococcus sp. strain with the ability of alkaline lipase secretion introduced by Slim et al.49 Additionally, alkaline lipase activity was observed in a Pseudomonas sp. strain, isolated from the peninsular coast of India.50 However, same ability was not detected for our Pseudomonas sp. K80. Also, extracellular alkaline lipase was purified from a new Bacillus sp. strain capable of withstanding pH 8.0 to 9.0.51 In the present study, same ability at pH 8.5 was observed mostly in the isolates of Halobacillus and Halomonas genera.
We found Bacillus and Halobacillus genera as potent extracellular pectinase producer in this study. CMCase activity was detected for the genus Halobacillus, Virgibacillus, Oceanobacillus, Staphylococcus, Pseudomonas, Idiomarina and Halomonas. In a survey of halophilic microorganism in Orissa and West Bengal, India, all halophilic bacteria were negative for pectinase and cellulose activities.52 In a screening of bacterial strains from Brazilian soil samples, 60.7% of isolated bacteria presented pectinolytic activity.35 The main microorganisms of Howz Soltan playa which were able to produce extracellular pectinase and cellulose belonged to the genus Gracilibacillus and Virgibacillus.41
Taxonomic Characterization of the Isolates
According to morphological (macro and microscopic) and molecular (sequences of genes encoding for 16SrRNA) analysis, only 21 isolates were selected and examined in greater details. Phylogenetic analysis of 16S rRNA sequences of the isolates revealed eight different taxonomic groups. Isolates clustered with Bacillus, Halobacillus, Virgibacillus, Oceanobacillus, Staphylococcus, Pseudomonas, Idiomarina and Halomonas with bootstrap values (24-100%). Figure 3 shows the phylogenetic tree of isolates. In contrast with the study of Rohban et al. the dominant genus of our study was Halobacillus and Halomonas they presented Salicola as the most frequent genus in Howz Soltan Lake.41 We found two members of genera Staphylococcus and Pseudomonas which previous studies have not reported. In the study of microbial diversity of Qinghai Lake, China, Actinobacteria and Acidobacteria/Holophaga was reported as the dominant gram-positive bacteria while we have not found them among our isolates.53 The most common bacteria from different saline environments in South Spain were assigned to genera Salinivibrio while we had not found them in our study.44 Although species of the genus Salinivibrio can be found in low salt concentration, in this study we did not identify any member of this genus.41 The present study as well as study of an alkaliphilic soda lake showed the most frequent isolates belonged to genera Halomonas and Bacillus.54
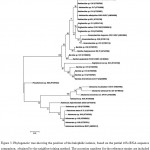 |
Figure 3. Phylogenetic tree showing the position of the halophilic isolates, based on the partial 16S rRNA sequence comparison, obtained by the neighbor-joining method. The accession numbers for the reference strains are included in brackets. Bootstrap values are indicated on the branches
|
Conclusions
To our knowledge, this is the first microbiological study on biodiversity of halophilic bacteria in Iraq. Lake Razazah is subjected to drastic physico-chemical conditions including changes in salinity, temperatures and dryness which make it a study target for microbiologists.In the course of a program of screening for halophilic and halotolerant bacteria in Razazah Lake, 218 halophilic bacteria were isolated from soil, water and sludge samples.We found halophilic bacteria belong to eight genera including Bacillus, Halobacillus, Virgibacillus, Oceanobacillus, Staphylococcus, Pseudomonas, Idiomarina and Halomonas. Additionally, the potential of various industrial hydrolase productions by halophilic bacteria demonstrated in this study. The genus Halobacillus, Halomonas and Bacillus had members with the most ability to produce industrial extracellular enzymes. Further studies focusing on molecular and biochemical characteristics recommend in order estimating the best hydrolytic exoenzyme producers. A large part of the oil resources in Iraq located in high temperature and hypersaline sites which are contaminated by crude oil. Apart from all industrial applications, discovery of new microorganisms which are able to treat these contaminations would be a perfect solution in environmental cleanup.
Competing Interests
The authors declare that they have no competing interests.
References
- Cantrell S. A., Martinez L. C and Molina M. Characterization of fungi from hypersaline environments of solar salterns using morphological and molecular techniques. Mycological Research. 2006;110(8):962-970.
CrossRef - Vijayanand S., Hemapriya J., Selvin J., Kiran S. Biodiversity of Extremely Halophilic Bacterial Strains Isolatedfrom Solar Salterns of Tuticorin, Tamilnadu, India. International Journal of Water Resources and Environmental Sciences. 2012;1(1):01-07.
- Sh A. G., Jamadar Z. A. S.,Vinod P. S. S and Sulochana M. B. Molecular characterization and screening of halophiles for the production of biopolymers. European Journal of Biotechnology and Bioscience. 2016;4(2):32-36.
- Hedi A., Sadfi N., Fardeau M. L., et al., Studies on the Biodiversity of Halophilic Microorganisms Isolated from El-Djerid Salt Lake (Tunisia) under Aerobic Conditions.International Journal of Microbiology. 2009.
CrossRef - DasSarma S., Berquist B. R., Coker J. A., DasSarma P and Müller J. A. Post-genomics of the model haloarchaeon Halobacterium sp. NRC-1. Saline Systems. 2006;2(3).
- Reid I. N., Sparks W. B., Lubow S and McGrath M. Terrestrial models for extraterrestrial life: methanogens and halophiles at Martian temperatures. International Journal of Astrobiology. 2006;5(2):89-97.
CrossRef - López-López M. A., Yarza P., Richter M. et al., Extremely halophilic microbial communities in anaerobic sediments from a solar saltern. Environmental Microbiology Reports. 2010;2(2):258-271.
CrossRef - Ali I., Prasongsuk S and Akbar A. Hypersaline habitats and halophilic microorganisms. Maejo International Journal of Science and Technology. 2016;10(3):330-345.
- Borgne S. L., Paniagua D and Vazquez-Duhalt R. Biodegradation of organic pollutants by halophilic bacteria and archaea. Journal of Molecular Microbiology and Biotechnology. 2008;15(2-3):74-92.
CrossRef - Zhao B., Wang H., Mao X and Li R. Biodegradation of Phenanthrene by a Halophilic Bacterial Consortium under Aerobic Conditions. Current Microbiology. 2009;58(3):205-210.
CrossRef - Mellado M. E., Ventosa A. Biotechnological potential of moderately and extremely halophilic microorganisms. In: Barredo J. L (ed) Microorganisms for health care, food and enzyme production. Research Signpost. Kerala. 2003; 233-256.
- Ventosa A., Nieto J. J and Oren A. Biology of moderately halophilic aerobic bacteria. Microbiology and Molecular Biology Reviews. 1998;62(2):504-544.
- Oren A. Diversity of halophilic microorganisms: Environments, phylogeny, physiology, and applications. Journal of Industrial Microbiology and Biotechnology. 2002;28(1):56-63.
CrossRef - Irshad A.,Ahmad I and Kim S. B. Culturable diversity of halophilic bacteria in foreshore soils. Brazilian Journal of Microbiology. 2014;45(2):563-571.
CrossRef - Abdelkafi S., Ogata H., Barouh N., et al., Identification and biochemical characterization of a GDSL-motif carboxylester hydrolase from Carica papaya latex. Biochimica et Biophysica Acta. 2009;1791(11):1048-1056.
CrossRef - Dharmaraj S. Marine Streptomyces as a novel source of bioactive substances. World Journal of Microbiology and Biotechnology. 2010;26(12):2123-2139.
CrossRef - Hasan F., Shah A. A and Hameed A. Industrial applications of microbial lipases. Enzyme and Microbial Technology. 2006;39(2):235-251.
CrossRef - Lee H. K., Lee J. K., Kim M. J and Lee C. J. Immobilization of lipase on single walled carbon nanotubes in ionic liquid. Bulletin- Korean Chemical Society. 2010;31(3):650-652.
CrossRef - Kamekura M. Production and function of enzymes of eubacterial halophiles. FEMS Microbiology Letters. 1986;39(1-2):145-150.
CrossRef - Onishi H and Sonoda K. Purification and Some Properties of an Extracellular Amylase from a Moderate Halophile, Micrococcus halobius. Applied and Environmental Microbiology. 1979;38(4):616-620.
- Shekha Y. A., Ismael H. M and Ahmed A. A. Bacteriological and Mycological Assessment for Water Quality of Duhok Reservoir, Iraq. Jordan Journal of Biological Sciences. 2013;6(4):308-315.
CrossRef - Al-mezori H. A and Hawrami K. A. M. Evaluation of Microbial qualiy of the drinking water of Duhok province/Kurdistanregion of Iraq. 2nd International Conference on Environmental Science and Development, IPCBEE. 2011;4 © (2011) IACSIT Press, Singapore. 2011.
- Salman J. M and Nasser A. J. Variation of some physico-chemical parameters and biodiversity of gastropods species in Euphrates River, Iraq. International Journal of Environmental Science and Development. 2013;5(3):328-331.
CrossRef - Kornijów R., Szczerbowski J. A., Krzywosz T and Bartel R. The macrozoobenthos of the Iraqi lakes Tharthar, Habbaniya and Razzazah. Archives of Polish Fisheries. 2001;9(1):127-145.
- Mohammad M. K. Existence and Ecology of the Burrowing Isopod Sphaeroma annandalei Stebbing. 1911. (Crustacea; Isopoda; Sphaeromatidae) in Lake Razzaza, Kerbala Province, Central Iraq. Advanced Biomedical Research. 2014;5(1):178-181.
- B. Zdanowski, K. Lossow, R. Bartel and J.A. Szczerbowski, “Salinity levels and the trophic state of Iraqi dam reservoirs and lakes. Archives of Polish Fisheries. 2001;9(l):35-52.
- Rahi K. A and Halihan T. Changes in the salinity of the Euphrates River system in Iraq. Regional Environmental Change. 2010;10(1):27-35.
CrossRef - Toma J. J. Limnological study of Dokan, Derbendikhan and Duhok lakes, Kurdistan region of Iraq. Open Journal of Ecology. 2013;3(1):23-29.
CrossRef - Smibert R. M and Krieg N. R. Edited by Gerhardt P.,Murray R. G. E.,Wood W. A and Krieg N. R. Phenotypic characterization. In Methods for General and Molecular Bacteriology, Washington, DC: American Society for Microbiology. 1994;607–654.
- Gregersen T. Rapid method for distinction of Gram-negativefrom Gram-positive bacteria. European journal of applied microbiology and biotechnology. 1978;5(2):123-127.
- Satellite Word Map. Available at http://satelliteworldmap.com/
- Pailin T., Kang D.H., Schmidt K and Fung D. Y. C. Detection of extracellular bound proteinase in EPS-producing lactic acid bacteria cultures on skim milk agar. Letters in Applied Microbiology. 2001;33(1):45-49.
CrossRef - Gonzalez C.,Gutierrez C and Ramirez C. Halobacterium vallismortis sp. nov. An amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Canadian Journal of Microbiology. 1978;24(6):710-715.
CrossRef - Zhou X., Chen H and Li Z. CMCase activity assay as a method for cellulose adsorption analysis. Enzyme and Microbial Technology. 2004;35(5):455-459.
CrossRef - Soares M. M. C. N., Silva R and Gomes E. Screening of bacterial strains for pectinolytic activity: characterization of the polygalacturonase produced by Bacillus sp. Revista de Microbiologia. 1999;30(4):299-303.
CrossRef - Tamura K., Stecher G., Peterson D., Filipski A and Kumar S. MEGA6: molecular evolutionary genetics analysis version 60. Molecular Biology Evolution. 2013; 30(12):2725-2729.
CrossRef - Saitou N and Nei M. The neighbor joining method a new method for reconstructing phylogenetic trees. Molecular Biology Evolution. 1987;4(4):406-425.
- Felsenstein J. Evolutionary trees from DNA sequences: amaximum likelihood approach.Journal of Molecular Evolution. 1981;17(6):368-376.
CrossRef - Fitch W. M. Toward defining the course of evolution: minimum change for a specific tree topology.Systematic Zoology. 1971;20(4):406-416.
CrossRef - Felsenstein J. Confidence limits on phylogenies: anapproach using bootstrap. Evolution. 1985;39(4):783-791.
CrossRef - Rohban R., Amoozegar M. A., Ventosa A. Screening and isolation of halophilic bacteria producing extracellular hydrolyses from Howz Soltan Lake, Iran. J Ind Microbiol Biotechnol. 2009;36(3):333-40.
CrossRef - Kumar S.,Karan R., Kapoor S., S. P. S and S. K. K. Screening and isolation of halophilic bacteria producing industrially important enzymes. Brazilian Journal of Microbiology. 2012;43(4):1595-1603.
CrossRef - Baati H.,Amdouni R., Gharsallah N., Sghir A and Ammar E. Isolation and characterization of moderately halophilic bacteria from Tunisian solar saltern. Current Microbiology. 2010;60(3):157-161.
CrossRef - Sánchez-Porro C., Martín S., Mellado E and Ventosa A. Diversity of moderately halophilic bacteria producing extracellular hydrolytic enzymes. Journal of Applied Microbiology. 2003;94(2):295-300.
CrossRef - de Souza P. M and de Magalhães P. O. Application of microbial α-amylase in industry – A review.Brazilian Journal of Microbiology. 2010;41(4):850-861.
CrossRef - Joo H. S and Chang C. S. Oxidant and SDS-stable alkaline protease from a halo-tolerant Bacillus clausii I-52: enhanced production and simple purification. Journal of Applied Microbiology. 2005;98(2):491-497.
- Patel R. K., Dodia M. S., Joshi R. H and Singh S. P. Production of Extracellular Halo-alkaline Protease from a Newly Isolated Haloalkaliphilic Bacillus sp. Isolated from Seawater in Western India. World Journal of Microbiology and Biotechnology. 2006;22(4):375-382.
CrossRef - Amoozegar M. A., Fatemia A. Z., Karbalaei-Heidarib H. R and Razavic M. R. Production of an extracellular alkaline metalloprotease from a newly isolated, moderately halophile, Salinivibrio sp. strain AF-2004. Microbiological Research. 2007;162(4):369-377.
CrossRef - Cherif S., Mnif S., Hadrich F., Abdelkafi S and Sayadi S. A newly high alkaline lipase an ideal choice for application in detergent formulations. Lipids in Health and Disease. 2011;10:221.
CrossRef - Kiran G. S., Shanmughapriya S., Jayalakshmi J., Selvin J., Gandhimathi R., Sivaramakrishnan S et al., Optimization of extracellular psychrophilic alkaline lipase produced by marine Pseudomonas sp. (MSI057). Bioprocess and Biosystems Engineering. 2008;31(5):483-492.
CrossRef - Sharma R., Sonia S. K., Vohrab R. M., Guptaa L. K and Gupta J. K. Purification and characterisation of a thermostable alkaline lipase from a new thermophilic Bacillus sp. RSJ-1. Process Biochemistry. 2002;37(10):1075-1084.
CrossRef - Biswas J and Paul A. K. Production of Extracellular Enzymes by Halophilic Bacteria Isolated from Solar Salterns. International Journal of Applied Biology and Pharmaceutical Technology. 2013;4(4):30-36.
- Dong H., Zhang G., Jiang H et al., Microbial Diversity in Sediments of Saline Qinghai Lake, China: Linking Geochemical Controls to Microbial Ecology. Microbial Ecology. 2006;51(1):65-82.
CrossRef - Duckworth A. W., Granta W. D., Jones B. E and van Steenbergen R. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiology Ecology. 1996;19(3):181-191.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





