How to Cite | Publication History | PlumX Article Matrix
Anita H. Permana1,4, Fida Madayanti Warganegara1, Deana Wahyuningrum2, Made Puspasari Widhiastuty1 and Akhmaloka1,3
1Biochemistry Research Group, Faculty of Mathematics and Natural Science, Institut Teknologi Bandung, Indonesia.
2Organic Chemistry Research Group, Faculty of Mathematics and Natural Science, Institut Teknologi Bandung, Indonesia.
3Department of Chemistry, Faculty of Science and Computer, Universitas Pertamina, Indonesia.
4Study Program of Quality Assurance of Food Industry, Politeknik AKA Bogor, Indonesia.
Corresponding Author E-mail: loka@chem.itb.ac.id
DOI : http://dx.doi.org/10.13005/bbra/2545
ABSTRACT: Heterologous expression and purification of thermostable lipase from Geobacillus thermoleovorans PPD2 had been carried out through Escherichia coli BL21 as host. Two bands obtained showed lipolytic activity with the size at around 51 (LipA) and 43 (LipB) kDa, respectively. The activities were identified by zymogram analysis, while the control protein from Escherichia coli BL21(DE3) do not show any lipolytic activity. Purification of crude extract using chromatography affinity Ni-NTA resulted one dominant band of LipA, meanwhile LipB did not appeared on the gel. Another purification for LipB was carried out by acetone fractionation. Both of LipA and LipB showed high activity toward medium chain length substrates, with optimum activity at 50oC and pH 8.5. The activities of LipA and LipB showed tolerance toward short chain alcohols, such as methanol, ethanol, n-propanol, and isopropanol.
KEYWORDS: Acetone Fractionation Geobacillus Thermoleovorans PPD2; Thermostable Lipase; Recombinant Protein; Ni-NTA Affinity Chromatography;
Download this article as:| Copy the following to cite this article: Permana A. H, Warganegara F. M, Wahyuningrum D, Widhiastuty M. P, Akhmaloka A. Heterologous Expression and Characterization of Thermostable Lipases from Geobacillus Thermoleovorans PPD2 Through Escherichia Coli. Biosci Biotech Res Asia 2017;14(3). |
| Copy the following to cite this URL: Permana A. H, Warganegara F. M, Wahyuningrum D, Widhiastuty M. P, Akhmaloka A. Heterologous Expression and Characterization of Thermostable Lipases from Geobacillus Thermoleovorans PPD2 Through Escherichia Coli. Biosci Biotech Res Asia 2017;14(3). Available from: https://www.biotech-asia.org/?p=27846 |
Introduction
Lipase is widely used in industrial processes; such as fat and fatty acids, organic synthesis, polymer, and bioenergy industries (Brígida, et al., 2014). The enzyme catalyzes both of hydrolytic and transesterification reactions, depends on the aqueous and organic composition in the solution. Thermostable lipases, as well as other thermostable enzymes, have some unique properties that would be useful for industrial application. Most of industrial processes are occurred at high temperature, which is the optimum temperature for thermostable enzymes. Other advantages of high temperature processes are the increased reaction rate, high solubility of substrate and reagent, efficiency in reactor usage, and minimum contamination of microorganism (Hasan, et al., 2006).
During the last five years, a lot of efforts have been carried out in order to produce active lipases using different producing microorganisms (Abdelmoez and Mustafa, 2014). Various sources of microorganism producing lipase have been explored, such as from hot spring (Febriani, et al., 2013; Shahinyan, et al., 2017; Sahoo et al., 2017), mud crater (Asy’ari, et al., 2014), petroleum sludge (Yılmaz and Sayar, 2015), and domestic compost (Syihab, et al., 2015; Nurhasanah, et al., 2015). Thermostable lipases from several thermophilic microorganisms have been identified.
Lipase from Thermus aquaticus, isolated from Indonesian hot spring, was obtained by homologous expression and purified by hydrophobic interactions-based chromatography. The enzyme showed as true lipase and had optimum activity at 75oC (Febriani, et al., 2013). Three isolates from Armenian geothermal springs, identified as Geobacillus sp., Bacillus licheniformis and Anoxibacillus flavithermus, also showed lipolytic activity on agar plates with olive oil emulsion (Shahinyan, et al., 2017).
Petroleum sludge and domestic compost showed promising results as a source for unique lipases. Cryptococcus diffluens D44 lipase, isolated from petroleum sludge (optimum activity at pH 9.0 and 45°C), showed increasing activity in methanol and some organic solvent (Yılmaz and Sayar, 2015). Syihab, et al. (2017) also reported that thermostable lipases from Pseudoxanthomonas sp., isolated from domestic compost, showed tolerance towards methanol and various organic solvent. The enzymes have optimum activity at alkaline condition (pH 8.0 and 10.0) and high temperature (50 and 70°C).
Another method to obtain thermostable lipase was carried out through metagenomic approach (Nurhasanah et al., 2015 and 2017; Sahoo et al., 2017). Five lipase genes, highly homolog to the lipase from Pseudomonas genus, have been cloned (Nurhasanah et al., 2015). Three of the genes were successfully sub-cloned, and the recombinant vectors were heterologically expressed into Escherichia coli BL21(DE3) (Nurhasanah et al., 2017). Heterologous expression of the lipase gene from metagenomics approarch, also reported by Sahoo et al. (2017). Optimum pH and temperature of purified lipase were 8.0 and 65°C. The enzyme could hydrolyse a wide range of 4-nitrophenyl esters, and resistant to many organic solvents.
Lipase gene from local thermophilic bacteria, isolated from Manuk Crater, has been cloned into Escherichia coli (Widhiastuty, et al., 2011). Heterologous expression of lipase from the Manuk isolate has been reported by Brilliantoro, et al. (2015); meanwhile the lipase from Papandayan isolate, Geobacillus thermoleovorans PPD2, has also been isolated in the previous research (personal communication). However, the expression of the gene was not carried out yet. In this paper, heterologous expression and characterization of the enzymes are reported.
Materials and Methods
Materials
G. thermoleovorans PPD2 isolate and its lipase gene were obtained from our collection. E. coli BL21(DE3)-pET-30a(+) and E. coli BL21(DE3) was used as a host cell. Luria Bertani medium (1% w/V Tryptone, 1% w/V Sodium chloride, and 0.5% w/V yeast extract), Kanamycin sulfate (Bio Basic), and IPTG (Thermo Scientific) were used for lipase expression. Polyacrylamide gel 10%, Spectra Multicolor Broad Range Protein Ladder (Thermo Scientific), and peqGOLD Protein Marker III (Peqlab) were used for protein electrophoresis. 1-naphtyl acetate (Sigma) and Fast Blue Dye (Bio Basic) were used for zymography analysis.
Ni-NTA Agarose (Qiagen, Germany), acetone (pro analysis), and Amicon Ultra-4 Centrifugal Filters (Merck Millipore) were used for protein purification. 2,4-nitrophenol (Sigma) was used as the standard for lipase activity assay. Various esters from 2,4-nitrophenol (palmitate, myristate, laurate, decanoate, butyrate, and acetate) were used as the substrate for lipase activity assay. Bradford reagent kit (Thermo Scientific) was used for determination of protein amount.
Methods
Heterologous Expression
E. coli BL21(DE3) harboring pITBlip2.2 plasmid was inoculated overnight (37oC, 150 rpm) in Luria Bertani (LB) medium which contains 0.1% kanamycin sulfate. The overnight culture was re-inoculated in fresh LB-kanamycin medium until the optical density (OD600) reached 0.60. Overexpression of recombinant lipase was conducted by induction with 1 mM IPTG, and the culture was shaken at 180 rpm within 4 hours. The culture was then subjected for centrifugation (6000 g, 30 minutes). The pellet was collected and suspended on lysis buffer (50 mM Na-phosphate buffer pH 8, 0.1% SDS), continued by 8 minutes boiling at 95oC. Protein crude extract was obtained by centrifugation (12000 g, 30 minutes) to separate the lysate from cell debris.
Affinity Chromatography
Affinity chromatography (using Ni-NTA Agarose matrix) included three steps: protein binding, washing and elution were used to purify the protein. 1.5 mL suspension of Ni-NTA Agarose was pipetted into column, and the resin was slowly separated from the solvent. The resin was equilibrated by 2 x 6 mL miliQ water, then followed by 3 x 6 mL binding buffer (50 mM K-phosphate buffer pH 8, 100 mM NaCl, 1% (V/V) Triton-X 100). Protein crude extract was then pipetted into the column, and the unbound proteins were collected as flowthrough. Furthermore, the column was washed with washing buffer (50 mM K-phosphate buffer pH 8, 100 mM NaCl). In the last step, the protein was eluted with elution buffer (50 mM K-phosphate buffer pH 8, 300 mM NaCl, imidazoles 10-200 mM). The crude extract, flowthrough, and elution fractions were subjected to diafiltration treatment using Amicon Ultra-4 Centrifugal Filters (Merck Millipore) to remove the remaining SDS and imidazole.
Acetone Fractionation
The unbound protein from the previous step (flowthrough fraction) was precipitated by acetone, and separated into three fractions: 0-40%, 40-60%, and 60-95% of acetone. The acetone was cooled at -20 oC within two hours before added to the protein. The protein-acetone mixture, for each fraction, was kept at 4 oC and then centrifuged at 12000 g for 20 minutes. The precipitate was then collected and resuspended in 50 mM K-phosphate buffer pH 8.
SDS-PAGE and Zymography
SDS-PAGE was performed on 10% running gels, and the bands were visualized by Coomassie Brilliat Blue staining. Gels to be stained for lipase activity was treated to a renaturation process in 50 mM buffer K-phospate pH 8 (containing 1% Triton X-100). The gel was incubated overnight at 4 oC, before it was stained for esterase/lipase activity in developing solution 1-naphthyl acetate as substrate (Soliman, et al., 2007).
Lipase Activity Assay
The activity assay was conducted with spectrophotometry analysis. One unit of lipase was defined as one μmol 2,4-nitrophenol released within a minute, while the specific activity was defined in unit per mg protein. Hydrolytic activity during the purification process was measured using 2,4-nitrophenol palmitate as the substrate. The activity was determined at 70 oC and pH 9.0, using the method described by Lee, et al. (1999) with some modification. The protein amount was determined using Bradford method (Bradford, 1976).
Substrate Specificity
The active fractions, which contain either LipA or LipB, was subjected to the assay using various 2,4-nitrophenol esters as the substrate: palmitate (C16), myristate (C14), laurate (C12), decanoate (C10), butyrate (C4), and acetate (C2). The assay was carried out at 70oC and pH 9.0.
Optimum Temperature and pH
Hydrolytic activity was measured at various temperatures (30, 40, 50, 60, 70, and 80oC) and pH (6.5, 7.0, 7.5, 8.0, 8.5, 9.0, and 9.5). Optimum pH was determined at 50oC, using K-phosphate buffer (pH 6.5 to 8.0) and Glysin-NaOH buffer (pH 8.5 to 9.5).
Organic Solvent Tolerance
The activity was measured in the presence of various organic solvent: methanol, ethanol, n-propanol, n-butanol, isopropanol, isoamyl alcohol, acetonitrile, acetone, chloroform, and n-hexane. The activity was determined at 50oC, pH 8.5, and 3% of organic solvent concentration.
Results and Disscussion
Expression and Purification of Recombinant Lipase
Heterologous expression of lipase gene in E. coli BL21(DE3) was carried out by induction of 1 mM IPTG. SDS-PAGE analysis showed two protein bands were expressed by the E. coli BL21(DE3)-pITBlip2.2 carrying recombinant plasmid, however the bands did not appear in the E. coli control (BL21(DE3)-pET-30a(+)) (Figure 1A). Both of the bands, with the sized of 51 (LipA) and 43 (LipB) kDa, showed lipase activity in the assay. The other bands, which represents protein from E. coli BL21(DE3)-pET-30a(+), do not show any lipase activity (Figure 1B). Based on the in silico analysis, the approximate weight of the fusion protein is around 51 kDa (LipA). It is confirmed by the first band, which is highly expressed under IPTG induction and the expression was reduced without IPTG induction (Figure 1A).
![Figure 1: Profile SDS-PAGE (1A) and zymogram (1B) of the lysate: [M] Spectra Multicolor Broad Range Protein Ladder, [1] Protein expressed by.](https://www.biotech-asia.org/wp-content/uploads/2017/10/Vol_14_no3_Het_Ani_fig1-150x150.jpg) |
Figure 1: Profile SDS-PAGE (1A) and zymogram (1B) of the lysate: [M] Spectra Multicolor Broad Range Protein Ladder, [1] Protein expressed by.
|
E. coli BL21(DE3)-pET-30a(+), [2] Protein expressed by E. coli BL21(DE3)-pITBlip2.2, without IPTG induction, [3] Protein expressed by E. coli BL21(DE3)-pITBlip2.2, induced by 1 mM IPTG. There were two bands that showed lipase activity, which sized at about 51 (LipA) and 43 (LipB) kDa.
The appearance of 43 kDa protein showing lipolytic activity was surprising since the protein did not appeared on crude extract of the control E. coli BL21(DE3)-pET-30a(+). This was only possible when the recombinant protein was post-translationally modified. The LipA still contains a putative signal peptide, when the region was cleaved resulting 388 amino acid residues corresponded to the protein with molecular weight of 43 kDa (LipB). The reduction of molecular weight due to a signal peptide cleavage was also observed on lipase from G. thermoleovorans YN (Soliman et al., 2007). If the signal peptide cleaved, the 43 kDa protein (LipB) was lost of His-tag fusion.
Purified LipA was obtained by boiling lysis method followed by Ni-NTA affinity chromatography. LipA, which contains His-tag fusion, easily bound into the matrix; meanwhile LipB could not bound to the matrix. LipB was released into the flowthrough, suggested that LipB had lost its His-tag (Figure 2A). LipA was removed from the matrix by elution with imidazole, with the optimum concentration of 100 mM imidazole (data not shown).
Since LipB could be purified by Ni-NTA affinity chromatography, it was partially purified by acetone fractionation of the unbound protein (Figure 2A). LipB was successfully separated from LipA at 60-95% acetone fraction. Although the fraction contains several protein bands (Figure 2A), the zymogram showed that LipB was the only protein that had lipolytic activity. Unfortunately, high concentration of acetone had reduced some of the enzyme activity, as shown on the zymogram (Figure 2B).
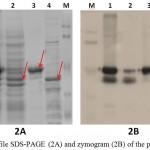 |
Figure 2: Profile SDS-PAGE (2A) and zymogram (2B) of the purified protein:
|
Specific activity of purified LipA and LipB was 68.5 ± 0.828 and 16.8 ± 0.151 U/mg protein, respectively. The specific activity of LipA was increased 11 fold compared to the respective crude extract, meanwhile LipB’s was 2.7 fold (Table 1). Both LipA and LipB have contribution to crude extract activity, and the purification process gave yield at 60.1% of LipA and 15.1% of LipB (Table 1). The results showed that most of the crude extract activity was preserved during the purification process.
Table 1: Purification of LipA and LipB. Unit activity (U): 1 µmol pNP released by the reaction per minute at 70oC and pH 9, using pNP palmitate as substrate.
| Sample | Volume (mL) | Total Protein (mg) | Total Activity (U) | Specific Activity
(U/mg protein) |
Fold | Activity Yield (%) |
| Crude extract | 15.5 | 29.7 ± 0.073 | 184.7 ± 2.42 | 6.21 ± 0.081 | 1 | 100 |
| LipA | 4 | 1.62 ± 0.002 | 111.0 ± 1.34 | 68.5 ± 0.828 | 11 | 60.1 |
| LipB | 1 | 1.71 ± 0.022 | 28.61 ± 0.26 | 16.8 ± 0.151 | 2.7 | 15.5 |
Optimum Temperature and pH
LipA and LipB showed similar pattern when the hydrolytic activities were measured at various temperatures and pH. Optimum activity for LipA and LipB were observed at 50oC. The enzymes maintained at least half of the activity at 70oC, but dramatically decreased at 80oC (Figure 3). On the variation of the pH, both LipA and LipB showed optimum activity at pH 8.5, indicated that the enzymes worked better at alkaline pH rather than acidic condition (Figure 4).
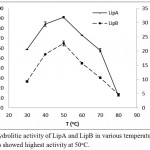 |
Figure 3: Hydrolitic activity of LipA and LipB in various temperatures. Both of the enzymes showed highest activity at 50oC.
|
The activity was dramatically decreased at 80oC, but the enzymes still maintained half of the activity at 70oC.
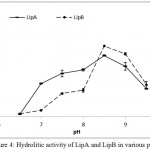 |
Figure 4: Hydrolitic activity of LipA and LipB in various pH.
|
Both of the enzymes showed highest activity at pH 8.5, indicated that the enzymes work better at alkaline pH.
Substrate and Solvent Preference
The fusion lipase (LipA) and possibly post-translational modification lipase (LipB) showed similar activity on the variation of chain length of the substrate. The activities were increased along with the reduction of chain length, and the highest activity was observed when the decanoate (C10) ester was used. The hydrolytic activities toward butyrate (C4) ester were dramatically decreased and lost when acetate (C2) ester was used (Figure 5).
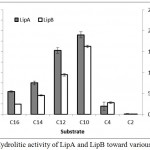 |
Figure 5: Hydrolitic activity of LipA and LipB toward various substrates.
|
The enzymes showed similar pattern, including high activity toward medium chain length substrates.
LipA and LipB still maintained the hydrolytic activity in sort chain length of alcohols, such as methanol, ethanol, n-propanol, and isopropanol (Figure 6). The presence of organic solvent offers high dissolution of hydrophobic compounds (Salihu and Alam, 2015), which were required to create an interface, for lipase activity (Willems et al., 2017). Recombinant lipase from Burkholderia cepacia G63 also reported stable in the presence of short chain alcohols, such as methanol, ethanol, t-butyl alcohol and glycerol (Yang et al., 2007).
When n-butanol and isoamyl alcohol were present, LipA and LipB showed dramatically loss of the activities (Figure 6). Longer hydrocarbon chain in the butyl and isoamyl groups reduced the polarity, resulted in inactivity of the enzyme. Huge activity loss was also observed on nonpolar solvent, such as n-hexane and chloroform. The results suggested that LipA and LipB most likely unstable in nonpolar solvent. Severas lipases obtained by homologous expression from Pseudomonas fluorescens P21 (Cardici and Yasa, 2010), Pseudoxanthomonas sp. (Syihab et al., 2017), and Bacillus sphaericus MTCC 7542 (Tamilarasan and Kumar, 2012), were reported to have wide range of organic solvent tolerance, from polar to nonpolar solvent.
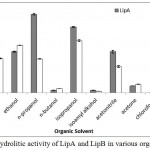 |
Figure 6: Hydrolitic activity of LipA and LipB in various organic solvent.
|
The enzymes still maintained their activities in the presence of acetonitrile and short chain length alcohol (C1-C3). Huge activity loss was observed in the presence of chloroform, n-hexane, and longer chain alcohols.
Lipase from Geobacillus sp. ARM, obtained by heterologous expression, had some activity retained in methanol, ethanol, and n-hexane; but the activity was inhibited in the presence of n-propanol, n-butanol, isopropanol, isoamyl alcohol, acetonitrile, and acetone (Ebrahimpour et al., 2011). Meanwhile, hydrolytic activity was drastically reduced in acetone (Figure 6). The characterization results showed no significant differences between Lip A and Lip B, this suggest that Lip B is the result of post translational modification of Lip A.
Acknowkledgement
We would like to thank to P3MI research project program of ITB, Ministry of Research, Technology and Higher Education to make this research possible to be carried out.
Conflict of Interest
The authors declare no conflict of interest that could appear to have influenced the submitted work.
References
- Abdelmoez W and Mustafa A. Oleochemical industry future through biotechnology. J. Oleo Sci. 2014;63(6):545-554.
CrossRef - Asy’ari M., Parwata I. P., Aditiawati P. A., Hertadi R. Isolation and identification of halostable lipase producing bacteria from the Bledug Kuwu mud crater located at Purwodadi-Grobogan, Central Java, Indonesia. J. Pure Appl. Microbiol. 2014;8(5):3387-3396.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72(1-2):248-254.
CrossRef - Brígida A. I., Amaral P. F., Coelho M. A., Goncalves L. R. Lipase from Yarrowia lipolytica: Production, characterization and application as an industrial biocatalyst. Journal of Molecular Catalysis B: Enzymatic. 2014;101: 148-158.
CrossRef - Brilliantoro R., Zidny R., Widhiastuty M. P., Akhmaloka. Hydrolytic and transesterification activities of ther mostable lipase ITB1. Biosciences Biotechnology Research Asia. 2015;12(1):01-06.
- Cadirci B. H., Yasa I. An organic solvents tolerant and thermotolerant lipase from Pseudomonas fluorescens P21. J Mol Catal B: Enzyme. 2010;64:155–161.
CrossRef - Ebrahimpour A., Rahman R. N. Z. R. A., Basri M., Salleh A. B. High level expression and characterization of a novel thermostable, organic solvent tolerant, 1, 3-regioselective lipase from Geobacillus sp. Strain ARM. Bioresource Technology. 2011;102(13):6972-6981.
CrossRef - Ihsanawati F., Hertadi R., Madayanti F., Akhmaloka. Thermostable alkaline lipase isolated from Thermus aquaticus. International Journal of Integrative Biology. 2013;14(2):104.
- Hasan F., Shah A. A., Hameed A. Industrial applications of microbial lipases. Enzyme and Microbial technology. 2006;39(2): 235-251.
CrossRef - Lee D. W., Koh Y. S., Kim K. J., Kim B.C., Choi H. J., Kim D. S., Suhartono M. T., Pyun Y. R. Isolation and characterization of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol. Lett. 1999;179(2):393-400.
CrossRef - Nurbaiti S. N., Warganegara F. M., Akhmaloka. Diversity of gene encoding thermostable lipase from compost based on metagenome analysis. International Journal of Integrative Biology. 2015;16(1):7.
- Nurbaiti S. N., Warganegara F. M., Akhmaloka. Heterologous expression of gene encoded thermostable lipase and lipolytic activity. J. Pure Appl. Microbiol. 2017;11(1):135-139.
CrossRef - Sahoo R. K., Kumar M., Sukla L. B., Subudhi E. Bioprospecting hot spring metagenome: lipase for the production of biodiesel. Environmental Science and Pollution Research. 2017;24(4):3802-3809.
CrossRef - Salihu A., Alam M. Z. Solvent tolerant lipases: a review. Process Biochemistry. 2015;50(1):86-96.
CrossRef - Shahinyan G., Margaryan A., Panosyan H., Trchounian A. Identification and sequence analyses of novel lipase encoding novel thermophillic bacilli isolated from Armenian geothermal springs. BMC microbiology. 2017;17(1):103.
CrossRef - Soliman N. A., Knoll M., Abdel-Fattah Y. R., Schmid R. D., Lange S. Molecular cloning and characterization of thermos table esterase and lipase from Geobacillus thermoleovorans YN isolated from desert soil in Egypt. Process Biochemistry. 2007;42:1090–1100.
CrossRef - Syihab S. F., Madayanti F., Akhmaloka. Isolation, characterization, and identification of lipolytic thermophiles with methanol tolerance from domestic compost. J. Pure Appl. Microbiol. 2015;9(2).
- Syihab S. F., Madayanti F., Akhmaloka A., Widhiastuty M. P. Purification and characterization of thermostable and alcohol tolerant lipase from Pseud oxanthomonas sp. African Journal of Biotechnology. 2017;16(31):1670-1677.
CrossRef - Tamilarasan K., Kumar M. D. Purification and characterization of solvent tolerant lipase from Bacillus sphaericus MTCC 7542. Biocatalysis and Agricultural Biotechnology. 2012;1(4):309-313.
CrossRef - Widhiastuty M. P., Febriani M., Moeis M. R., Akhmaloka., Madayanti F. Cloning, homological analysis and expression of lipase gene from hot spring isolate. International Journal of Integrative Biology. 2011;11(1):8-13.
- Willems N., Lelimousin M., Koldsø H., Sansom M. S. Interfacial activation of M37 lipase: A multi-scale simulation study. Biochimica et Biophysica Acta (BBA)-Biomembranes. 201;1859(3):340-349.
CrossRef - Yang J., Guo D., Yan Y. Cloning, expression and characterization of a novel thermal stable and short-chain alcohol tolerant lipase from Burkholderia cepacia strain G63. J. Mol. Catal. B: Enzyme. 2007;45:91-96.
CrossRef - Yılmaz D. E and Sayar N. A. Organic solvent stable lipase from Cryptococcus diffluens D44 isolated from petroleum sludge. Journal of Molecular Catalysis B: Enzymatic. 2015;122:72-79.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





