How to Cite | Publication History | PlumX Article Matrix
Vikas Kumar1, Neelkamal Sharma1, Kusum Singal1 and Arun Sharma2
1Department of Genetics (Forensic Science), Maharshi Dayanand University, Rohtak Haryana, India.
2Directorate of Forensic Science, Himachal Pradesh, Shimla Hills, Junga.
Corresponding Author E-mail: neelforensics@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2621
ABSTRACT: Identification of exhibits obtained in wildlife cases usually presents challenging tasks for the forensic science investigators. This study describes a casework, where a degraded tissue sample was recovered from pathology department LUVAS University Hisar, Haryana to resolve the identity of the questioned sample. The mitochondrial DNA region of the questioned sample was amplified and sequenced using universal primers of cytochrome b gene to determine the forensically informative nucleotide sites to find the species identity. The obtained sequencing results were compared with the most homologous sequences extracting from NCBI-GenBank database and a phylogenetic tree was done with the aligned sequences to determine the species identity with strong bootstrap support. The informative sites generated revealed that the degree of sequence similarity showed maximum homology (100%) with the sequence obtained from the database. Based on the FINS analysis the recovered sample related to Antilope cervicapra (family Bovidae).
KEYWORDS: Blackbuck Cytochrome b; Forensic Science;FINS;NCBI; Phylogenetic; Wildlife;
Download this article as:| Copy the following to cite this article: Kumar V, Sharma N, Singal K, Sharma A. A Wildlife Forensic Study for the Species Identification of Indian Blackbuck Through Forensically Informative Nucleotide Sequencing (FINS). Biosci Biotech Res Asia 2018;15(1). |
| Copy the following to cite this URL: Kumar V, Sharma N, Singal K, Sharma A. A Wildlife Forensic Study for the Species Identification of Indian Blackbuck Through Forensically Informative Nucleotide Sequencing (FINS). Biosci Biotech Res Asia 2018;15(1). Available from: https://www.biotech-asia.org/?p=29315 |
Introduction
Species Identification of different biological trace evidence is one of the prime tasks for wildlife forensic scientists. Wildlife forensic in itself a very broad and challenging discipline, which has to deal with a plethora of wildlife species to fight against wildlife crime.1 Lack of stringent legislation and adequate identification tools for the many of the wildlife species make the situation worst to check the wildlife crime.2 With the advent of molecular science and conservation genetics practices plays an important role to meet the increasing need of investigating tool for wildlife investigating agencies.3
Amongst different molecular techniques, forensically informative nucleotide sequencing (FINS) has been one of the most advocated and widely accepted methods for the species identification.4-6 In this technique, investigators use universal primers to target the highly conserved species-specific mitochondrial genes segment viz. cytochrome b (Cyt b), 12s rRNA, 16s rRNA, for the identification of the species origin.7,8 The Cyt b gene is the most frequently used and widely accepted for phylogenetic work and species identification. The Cyt b gene evolves slowly in terms of non-synonymous substitutions, the rate of evolution in silent positions is relatively fast and conserved enough for clarifying deeper phylogenetic relationships.9
FINS based identification technique basically comprises of five steps, namely (A) DNA isolation from the recovered biological samples (B) Amplification of the selected specific DNA segment using universal primers (C) Determine the sequence of the amplified DNA segment (D) Match the sequencing results with the sequence available on the DNA Database (NCBI-GenBank) (F) Phylogenetic analysis with the sequence extractive from the database.10 This scientific approach is popularized as ‘Forensically Informative Nucleotide Sequencing (FINS)’ and particularly useful for wildlife forensics to identify the species. Usually Cytochrome b gene is widely used for the analysis of the species-specific identification, phylogenetic and forensic investigations.11 The present study uses the partial sequence of cytochrome b gene for the identification of forensically informative sites and applied FINS technique for the identification of wildlife species.
Material and Methods
Case History
A tissue sample was collected from a highly degraded animal body from the pathology department LUVAS University, Hisar, where a wildlife case was received for the postmortem from the wildlife department of Haryana to resolve the identity of the species. Tissue sample was stored at -20°C.12
DNA Extraction, PCR Amplification, and Sequencing to Establish The Identity
DNA was extracted from the questioned sample, using DNeasy blood and tissue Kit (Qiagen, Valencia, CA, USA) following manufacturer’s protocol instruction with little modifications by enhancing cell lysis time to 2-4 hours. Universal primers of cytochrome b gene viz. mcb398 (Fwd 5′-taccatgaggacaaatatcattctg-3′) and mcb869 (Rev5′-cctcctagtttgttagggattgatcg-3′) were used for the PCR amplification of the desired segment.13 PCR amplification was performed in a reaction volume of 20µl containing reaction composition: 1X PCR Buffer , 2.5 mM MgCl2, 0.2 mM of each of dNTPs, 10 pM each of forward and reverse primers, one units of Taq polymerase (Promega Biotech India Pvt. Ltd) and 30–40 ng of purified genomic DNA.
PCR Amplification was carried out for an initial 5 min at 94°C followed by 30 cycles at 94°C, 58°C, each for 45 sec and 72°C for 75 sec, and a final elongation step for 10 min at 72°C in a Takara PCR System 9700. 5µl of amplified PCR products were subjected and confirmed in 1.5% agarose gel stained with ethidium bromide and visualized gel doc system to detect the amplification. These amplified PCR products were sequenced (Eurofins Genomics India Pvt. Ltd) on both strands and results were checked, analyzed and edited in Chromas 2.4 (Technelysium Pty Ltd., South Brisbane, Australia).
Data Analysis
The obtained partial sequence data of the Cytochrome b gene were blasted for similarity search against non-redundant sequence database using BLAST tool of NCBI database (National Center for Biotechnology Information-Basic Local Alignment Search Tool).14-16 Most homologous matched sequences along with the closely related species were retrieved from NCBI-GenBank database and aligned with the sequence obtained from the questioned sample using MUSCLE program in Mega 7 software. Table 1 showing the species sequences from NCBI GenBank database used for comparison along with their accession numbers.17
Table 1: Showing the list of specie specific sequences of cytochrome b gene from GenBank database used for comparision.
| GenBank accession No |
| KU560641.1 (Antilope cervicapra) KU560640.1(Antilope cervicapra), JN632598.1(Antilope cervicapra), HQ122572.1(Antilope cervicapra), | AP003422.1(Antilope cervicapra), AF540921.1(Antilope cervicapra), AF034723.1(Gazella granti), JN632666.1(Nanger granti), JF728775.1 (Nanger dama), JF728776.1(Nanger soemmerringii), JN632667.1(Nanger soemmerringii) |
Results and Discussion
Sequential analysis showing highest sequence similarity in NCBI-GenBank database with the Blackbuck sequences. The following Cytochrome b sequence of the question sample was used for conducting the analysis given as follows:
TGAGGAGCAACAGTCATCACCAATCTCCTTTCAGCAATCCCATACATCGGTACAAACCTAGTAGAATGAATTTGAGGCGGGTTCTCAGTAGATAAAGCAACACTCACCCGATTTTTTGCCTTCCA CTTTATCCTCCCATTTATCATTGCAGCCCTTGCCATAGTTCACCTACTATTCCTTCACGAAACAGGATCCAACAACCCCACAGGAATTTCATCAGACGCAGACAAAATTCCATTCCACCCCTACTA CACTATCAAAGATATCCTAGGAGCTCTACTATTAATTTTAACCCTCATGCTTCTAGTCCTATTCTCACCGGACCTGCTTGGAGACCCAGACAACTATACACCAGCAAACCCACTTAATACACCCCCAC
The aligned sequences were subjected to the phylogenetic analysis using the Neighbour-Joining method, based on Kimura-2-parameter distance matrix (K2P) model with 1000 bootstrap replication, generated in MEGA v7.0 software.18-19 The NJ phylogenetic tree showed that the source of the study sample originates from the Blackbuck species (shown in Figure 1). After comparison and confirmation from the database, results were again confirmed by matching the obtained DNA sequence with the sequence of the reference sample of Blackbuck (result not shown). Blood sample as a reference sample was collected from the wildlife department during a veterinary checkup camp from Deer park Hisar, Haryana. The sequence of the reference sample for cytochrome b gene has submitted to the GenBank/NCBI database (Accession number MF434527).
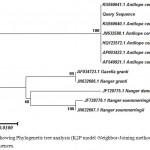 |
Figure 1: Showing Phylogenetic tree analysis (K2P model -Neighbor-Joining method) using Cyt b gene sequences.
|
Conclusion
On the basis of our findings, we clearly conclude that the forensic sample belongs to Blackbuck (Antilope cervicapra) which is a Schedule I animal, protected under the Wildlife (Protection) Act 1972, Government of India. Our results show the potential of FINS technology in wildlife forensics and also highlights the importance as a reliable technique in species identification even from highly degraded samples which is not possible sometimes by conventional means. Further, most of the species-specific gene sequences are not available in the database or most of the available sequences for particular species are either incomplete or partial. In such circumstances, reference sequences of the known species is the only way for the cross confirmation of the results. The success of FINS technique highly depends upon the richness of the database and this is possible when a stronger DNA database available for different species around the world to assist DNA based identification related to wildlife-crimes.
Acknowledgments
The authors are grateful to the Department of MOEF &CC (Wildlife Division), Government of India and Department of Forest (Wildlife Division) Haryana, for giving permission for sample collection. We also thanks to the staff of the pathology department LUVAS University, Hisar for providing postmortem samples. A sincere thanks to University Grant Commission for providing Junior Research Fellowship, the financial support is highly acknowledged.
Ethics of human and animal experimentation
Institutional Animal Ethics Committee (IAEC) was approved this research work vide letter no 151–67/30–03-2015. Sample collection permission for this scientific research is granted by the Ministry of Environment & climate changes (MoEF&CC) government of India and state Forest & Wildlife department Haryana vide letter no 1–56/2016 and WL-87/11–04-16 respectively.
Conflict of interest
There is no conflict of interests.
Reference
- Iyengar A. Forensic DNA analysis for animal protection and biodiversity conservation: A review. Nat. Conser. 2014;22(3):195-05.
CrossRef - Thakur M, Singh S.K, Shukla M, Sharma L.K, Mohan N. Identification of Galliformes through Forensically Informative Nucleotide Sequencing (FINS) and its Implication in Wildlife Forensics. Forensic Res. 2013;3:1-5.
- Cooper J.E, Cooper M.E. Wildlife forensic investigation: principles and practice. CRC Press, UK.
- Guha S, Kashyap V.K. Molecular identification of lizard by RAPD & FINS of mitochondrial 16s rRNA gene. Med. 2006;8(1):5-10.
CrossRef - Li M, Zhang K.Y.B, But P.P.H, Shaw P.C. Forensically informative nucleotide sequencing (FINS) for the authentication of Chinese medicinal materials. Med. J. 2011;6(1):42.
CrossRef - Rajpoot A, Kumar V, Bahuguna P, Kumar A. Forensically informative nucleotide sequencing (FINS) for the first time authentication of Indian Varanus species: implication in wildlife forensics and conservation. Mitochondrial DNA Part A .2016. doi.org/10.1080/24701394.2016.1202943.
CrossRef - Linacre A, Tobe S.S. An overview to the investigative approach to species testing in wildlife forensic science. Genet. 2011;2:2-9.
CrossRef - Sahajpal V, Goyal S.P. Identification of a forensic case using microscopy and forensically informative nucleotide sequencing (FINS): A case study of small Indian civet (Viverricula indica). Justice. 2010;50(2):94-7.
CrossRef - Bhaskar R, Khan I, Goyal S.P. Identification of forensic case using molecular markers: A case study of hyaena (Hyaena Hyaena). J. Pharma. Bio. Sci. 2011;2(4):561-71.
- Bartlett S.E, Davidson W.S. FINS (forensically informative nucleotide sequencing): a procedure for identifying the animal origin of biological specimens. Biotechniques. 1992;12(3):408-11.
- Parson W, Pegoraro K, Niederstatter H, Foger M, Steinlechner M. Species identification by means of the cytochrome b gene. J. Legal. Med. 2000;114(1):23-8.
CrossRef - Kumar N.V. Wildlife DNA Evidence: Recognition, Collection and Preservation. J. Forensic. Sci. 2015;3(7):8-15.
- Verma S.K, Singh L. Novel universal primers establish identity of an enormous number of animal species for forensic application. Ecol. Resour. 2003;3(1):28-31.
- Kumar V, Sharma N.A. DNA barcoding as a wildlife forensic tool (WFT) for species identification: a case study on Blackbuck (Antilope cervicapra) in India. J. Adv. Res. 2017;56:1283-87.
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden T.L. NCBI BLAST: a better web interface. Acids. Res. 2008;36(2):5-9.
CrossRef - Benson D.A, Cavanaugh M, Clark K, Karsch-Mizrach I, Lipman D.J, Ostell J, Sayers E.W. GenBank. Acids. Res. 2012;41(1):36-42.
CrossRef - Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Biol. Evol. 2011;28(10):2731-39.
CrossRef - Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Biol. Evol. 2016;33(7):1870-74.
CrossRef - Kumar V, Sharma N, Sharma A. DNA barcoding of the Indian blackbuck (Antilope cervicapra) and their correlation with other closely related species. Egypt. J. Forensic. Sci. 2017:7(1):31-8.

This work is licensed under a Creative Commons Attribution 4.0 International License.





