How to Cite | Publication History | PlumX Article Matrix
Intra-Cultivar Variability Endorsed by SSR Markers in Mango
Anju Bajpai1 , Nimisha Sharma2
, Nimisha Sharma2 , Navin Srivastava1,3
, Navin Srivastava1,3 , Shailendra Rajan1
, Shailendra Rajan1 and Muthukumar M1
and Muthukumar M1
1ICAR-Central Institute for Subtropical Horticulture, Lucknow, Uttar Pradesh 226101, India.
2ICAR-Indian Agricultural Research Institute, New Delhi 110012, India.
3Datar Genetics Limited, Nasik, Maharashtra, India.
Corresponding Author E-mail: muthukumarbt@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2622
ABSTRACT: Appreciable intra cultivar variation has warranted clonal selections, which has emerged as an important tool in mango breeding. This important source of morphological variability manifested in altered fruit and quality attributes has yielded improved clones in India and abroad, few of which were exposed to SSR based analysis. Statistical parameters viz., Polymorphic Information Content (0.319 in MiIIHR12 to 0.819 for MiIIHR26) and Gene Diversity (0.399 in MiIIHR12 to 0.839 for MiIIHR26), defined the ability of the chosen SSR markers to discriminate the intra cultivar variability, besides highlighting the extent of diversity captured by the improved clones. Furthermore, genetic relationship among the clones derived by Wards minimum variance, placed Himsagar and Langra clones in Cluster I and II respectively, Himsagar recording high genetic heterogeneity within its cluster, intra cultivar variability being 0.16-0.916, thus showing suitability for breeding by clonal selections. Even the use of limited SSR marker loci (6), could reveal and document the genomic variations accounting for the variations in the Dashehari clones as well as placing land race ‘Suraiyya’, as an out-group. The sampled clones of elite variety Chausa did not show any variation at the studied marker loci, thus exposing limited heterogeneity in the clones and demanding more explorations for breeding superior types targeting regularity in bearing.
KEYWORDS: Allelic Diversity; Dashehari; Genetic Variability; Heterogeneity; Himsagar
Download this article as:| Copy the following to cite this article: Bajpai A, Sharma N, Srivastava N, Rajan S, Muthukumar M. Intra-Cultivar Variability Endorsed by SSR Markers in Mango. Biosci Biotech Res Asia 2018;15(1). |
| Copy the following to cite this URL: Bajpai A, Sharma N, Srivastava N, Rajan S, Muthukumar M. Intra-Cultivar Variability Endorsed by SSR Markers in Mango. Biosci Biotech Res Asia 2018;15(1). Available from: https://www.biotech-asia.org/?p=29533 |
Introduction
Mango (Mangifera indica L.) holds pride in the Indian palette owing to its delicious taste, intimate association with history of its agriculture and civilization, well documented in religious scriptures, historical travelogues and architectural monuments dating back to more than 4,000 years (Prakash and Khan, 2005). Despite enormous genetic diversity, only few cultivars viz., Dashehari, Langra, Chausa, Banganapalli, Alphonso, Kesar and Himsagar and few others, dominates > 60 per cent of mango production area in India (Negi, 2000). Exploitation of natural variability through selection of superior clones of mango cultivar has been done by several workers. Naik (1948) observed significant variation among the trees of same clones in an orchard with respect to fruit shape, colour and quality which was ascribed to bud mutations. Oppenheimer (1956) reported existence of a wider variability in the performance of the trees belonging to same variety in same orchard, after surveying many orchards in India. Significant variation in fruit shape, size, colour and quality, possibly due to bud mutation is reported in “Alphonso” (Manchekar et al. 2011), most exported variety from Karnataka and Maharashtra. Similarly clonal selections have led to identification of superior clones viz. “Dashehari 51” (Negi, 1997) from Lucknow, “Banarasi Langra” (Singh et al. 1985) “Cardozo Mankurad” (Mathew and Dhandhar, 1997) from different locations in India. High yielding clones of Langra (Singh et al. 1985) and Kensington (Whiley et al. 1993) with resistance to black spot are also mentioned in literature.
In perennial trees like mango, clonal propagation coupled with traditional clonal selection methods have proven to be most efficient methods for capturing genetic potential including dominance, additive and epistatic interactions (Janick, 2006). Interestingly in all these cases the clonal multiplication is dependent on vegetative multiplication rather than true clonal (apomictic) multiplication. Clonal selection for high yield and quality led to collection of 50 clones from Malihabad, Rahimabad and Kakori areas of mango growing belts in Lucknow (Chadha et al. 1993). Till date, limited reports on molecular markers to assess clonal variability are available for grapes, apple and mango (Vignani et al.1996; Pancaldi et al.1998; Singh et al. 2009). The present study is the first report that was aimed to capture the genetic polymorphism using SSR markers and assess the clonal diversity of Dashehari, Langra, Chausa and Himsagar cultivars. The major focus of the investigation were (1) to demonstrate the resolving power of SSRs for evaluation of intra varietal clonal variability; (2) to investigate the genetic diversity within and among clonal populations of these for assessing the extent of inherent variability that can be suitably exploited towards varietal development.
Materials and Methods
A total of 24 mango clones from the Field Gene Bank at the ICAR-Central Institute for Subtropical Horticulture (CISH), Lucknow and Mohanpur, West Bengal, were used in the study. Total genomic DNA was extracted from fresh, young and healthy leaves as described by Dellaporta method (1983) with slight modifications as described in SSR genotyping of mango cultivars by Bajpai et al. (2016). The quality was checked on agarose gel (0.8%) and quantified using UV-spectrophotometer (Chemito).
Twenty SSR loci were employed for genotyping of mango clones. Polymerase chain reaction (PCR) amplification was performed using fluorescently- labeled primers with 5’-FAM or 5’-HEX using protocol as described by Ravishankar et al. (2011). The PCR products were checked for amplification on 3% agarose gel, further analyzed by gene scan analysis using ABI PRISM 310 automated fluorescent sequencer (GeneScan 3.1 analysis) and their allele sizes were determined.
The allelic composition of each accession and the number of total alleles were determined for each SSR locus, putative alleles were indicated by the estimated size in bp. The major allele frequency, polymorphism information content and gene diversity were calculated using Power Marker v3.25 (Liu and Muse, 2004). Genetic relationships among the genotypes were calculated by computing the dissimilarities through simple matching coefficient. A dendrogram was constructed through Wards Minimum Variance method using DARWin 5 software (Perrier et al. 2003).
Results and Discussion
Out of 20 SSR primers screened, 11 primers produced reproducible amplicons out of which six loci (Table- 1) were genotyped using Capillary electrophoresis for precise and accurate allele calling. In general, fluorescent-based analysis revealed single main peak in homozygotes and two different sized allelic peaks in heterozygotes. All these 6 microsatellites detected polymorphism following a disomic i.e. we observed a maximal occurrence of 2 alleles per locus. A range of 2-8 alleles with a cumulative allelic number 31 and a mean allele number of 5.16 in the size ranges 98 to 241 bp were recorded. The individuals used in the study represented superior clones of well established varieties from North and Eastern states of India and the intra varietal allelic variation among the closely related clones could be accounted by INDELs or single base variations. The fluorescent approach offered greater clarity of results interpretation as compared to silver stain approach as far as allele sizing and differentiation is concerned (Figure 1). Based on allelic diversity and polymorphism, MiIIHR26 was found most informative (Figure 1). PIC values ranged from 0.3197 to 0.8193 (average 0.636). Genetic variation estimated by heterozygosity (proportion of individuals heterozygous at a locus) was maximum for MiIIHR13 and allelic diversity (8) for MiIIHR26, respectively. The gene diversity denoted by expected heterozygosity (He) varied from 0.399 (MiIIHR12) to 0.839 (MiIIHR26), with an average value of 0.682. These characteristics summarize cumulative variations present in the clones of the commercial cultivars.
Table 1: SSR loci and primer details including allele size and polymorphism
| Locus | Primers(5’-3’) | Allele size (bp) | Allele no. | Major Allele Frequency | Gene Diversity | He | PIC |
| MiIIHR26 | F: GCGAAAGAGGAGAGTGCAAG
R: TATAAGTGCCCCCTCACG |
130-157 | 8 | 0.206 | 0.839 | 0.793 | 0.819 |
| MiIIHR32 | F: TGGTGGTGTTTGTTTGCAGT
R: ACCACCCGCAGTATTGAAAG |
182-194 | 4 | 0.396 | 0.722 | 0.827 | 0.678 |
| MiIIHR13 | F: CCCAGTTCCAACATCATCAG
R: TTCCTCTGGAAGAGGGAAGA |
182-199 | 4 | 0.362 | 0.695 | 0.931 | 0.635 |
| MiIIHR36 | F: TCTATAAGTGCCCCCTCACG
R: ACTGCCACCGTGGAAAGTAG |
221-241 | 5 | 0.258 | 0.770 | 0.551 | 0.731 |
| MiIIHR28 | F: AGCGGTCGCAGACAAATTCTATAT
R: ACAACTCGAGATTGTCACATCTTT |
98-118 | 8 | 0.517 | 0.667 | 0.758 | 0.630 |
| MiIIHR12 | F: GCCCCATCAATACGATTGTC
R: ATTTCCCACCATTGTCGTTG |
169-175 | 2 | 0.724 | 0.399 | 0.137 | 0.319 |
| TOTAL | 0.410 | 0.682 | 0.666 | 0.636 |
*F = forward and R = reverse, PIC – Polymorphism information content, He-Expected heterozygosity
 |
Figure 1: Agarose gel profile of Dashehari (A1) and Chausa (A2) clones at MiIIHR26 and MiIIHR13 loci, respectively (M-100bp marker) ;(B) Electrophoretogram of Himsagar clone 3 and 4 at MiIIHR26 locus displaying intra cultivar variation.
|
Ward’s minimum variance was used to determine genetic relationships among the sampled clones of three varieties and Langra types (Figure 2). Intra varietal variability described by genetic dissimilarity ranged from nil in case of Chausa clones, to 0 to 0.16 in case of Dashehari clones, while it was noticeably high in Himsagar clones (0.16 to 0.916). Dissimilarity matrix was further used to construct the dendrogram which placed the clones in two broad groups, between cluster distances averaging 0.575. The first group depicted more heterogeneity in the clones arranging 5 Himsagar clones with average dissimilarity of 0.525. The heterogeneous group of Himsagar clones offers wide scope of clonal selection as within variety variation is almost equaling variations recorded by comparing two elite North Indian varieties Dashehari and Chausa clones, as described earlier. Among the three Langra types that were studied, it is too premature to assume any relationship as they differ widely in morphology and can be source of detailed analyses. The second group is further divided into two (cluster III and IV), discreetly placing Dashehari and Chausa clones. The discriminatory potential of SSR markers was clear with Dashehari clones, as one mislabeled clone Dashehari C3, i.e., a superior mango landrace Surraiya, was placed as an out-group. Thus, SSR markers were not only able to confirm genetic similarity in Dashehari and Chausa clones, and variability in Himsagar clones, they could also correctly label C3 as an out group in Dashehari types. The study was able to demonstrate the ability of Gene Scan 3.1 analysis for efficient SSR allele discrimination at closely related clonal populations.
Role of genetic mutations as the basis of clonal differences has been long accepted. In grapes, particularly, owing to discovery of graft-transmissible diseases, rootstocks and ecology are also considered crucial for trait expression in plants of same cultivar. Genetic basis of clonal variations was proven irrevocably with tissue culture technology (Mannin et al. 1997). Even though the planting material is multiplied vegetatively in mango (softwood grafting or epicotyl grafting), often clonal variation is routinely observed under field conditions (Patil et al. 2006; Prasanth et al. 2007). Conceptually, mango clone individuality relies on propagation records and morphological features leading to breeding concept of clonal selection, particularly well documented in stone fruits (Gradziel, 2012). Accordingly, the selected clones of three important commercial varieties viz., Dashehari, Chausa and Himsagar as well as three types of Langra variety, cumulatively accounting for a population of 24 mango samples exposed to SSR genotyping yielded interesting observations. The molecular basis behind the generation of sports in woody perennial plants is not well documented. Genotyping data for the 6 SSR loci, across the clones of the 4 varieties, yielded allelic information in band sizes. Interestingly 10 Dashehari clones were clustered together, only C3 forming out group, indicating role of genomic marker for its differentiation. All 5 clones of Chausa were detected uniform genotypically at the studied loci. Langra type 2 was a variant among 3 clones studied, differing from other two selections at 3 positions. Altogether the population of Himsagar clones was most heterogeneous in four of sampled 6 loci, clone 4 having variation in each (Figure- 2).
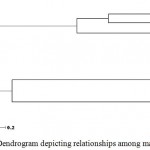 |
Figure 2: UPGMA Dendrogram depicting relationships among mango clones
|
The data suggests interaction of environment and somatic mutations or bud sport as progenitors of the variant Himsagar types, and also mislabeling of Himsagar seedlings. Earlier, similar studies in grapevine cv. ‘White Riesling’ clones Renger et al. (2000) established superiority of SSRs over other marker classes and proved that genetic mutations are the basis of clonal differences in grapes. The material also offers unique opportunities of studying epigenetic interactions in Dashehari and Chausa clones where altered phenotypes in identical clone (genotype) in same environment could be assigned to epigenetic rather than genetic factors (Janick, 2006). Desirable mutations in otherwise superior commercial varieties are likely to succeed due to discreet variations in otherwise superior genetic background. Presence of retrotransposon like elements could also be ascribed to clonal variability in mango as reported in Citrus (Liu et al. 2009). The role of clonal selection in mango has been studied in India and abroad (Singh and Chadha, 1981; Mukherjee et al. 1985; Whiley et al. 1993). Variations within clones of known varieties are reportedly due to rootstock and scion interaction in perennial tree species (Hartmann and Kester, 2002). Perpetuation and fixation of bud mutation is facilitated in obligate out crosser like mango, as favorable heterozygosity is conserved by clonal multiplication (Mckey et al. 2010). As genetic diversity in clonally propagated crops is eroded if no mechanism of generates new diversity, variations exhibited by improved clones at morphological and molecular level are drivers for the variability in clonal populations. An interesting ecological insight reveals that somatic mutations provide necessary genetic variation contributing to adaptive evolution (Whitham and Slobodchikoff, 1981) and if inherited by gametes, a source of variation equivalent to mutations during meiosis (Orive, 2001). In summary, maintaining adaptive potential of mango over varied agro climates is dependent on their capacity to generate diversity. Some clonal selection in elite varieties of Northern Indian plains assessed by molecular analysis using SSR markers detected polymorphism among the varieties and utilized for genetic relationship analysis. Based on molecular data, Himsagar clones were found to exhibit heterogeneity, thus showing suitability for further selections. The sampled clones of elite variety Chausa, did not show any variation at the studied marker loci, demanding more selections for its improvement.
Conclusions
Intra cultivar variation, an important source of morphological variability manifested in altered fruit and quality attributes, exposed to SSR marker analysis, revealed genetic nature of these variations. Dashehari clones revealed limited genetic variability (0-0.16); ‘Suraiyya’ being mislabeled as Dasheheri clone (C3), yet Himsagar clones were found to exhibit heterogeneity, thus showing suitability for further clonal selections. In summary, core of clonal variation displayed by morphological and molecular variations, was captured very well by SSR markers in mango.
Acknowledgements
Authors acknowledge Director ICAR-CISH and the Department of Biotechnology, Government of India for providing budgetary support.
References
- Bajpai A, Muthukumar M, Ahmad I, Ravishankar K.V, Parthasarthy V.A, Sthapit B, Rao R, Verma J.P, Rajan S. Molecular and morphological diversity in locally grown non-commercial (heirloom) mango varieties of North India. J. Environ. Biol. 2016;37(2):221-228.
- Chadha K.L, Yadav I.S, Sinha G.C, Rajan S, Singh H. Clonal selection in mango for high yield and quality (Abs.) In: Golden Jubilee Symposium Horticultural Research-Changing Scenario at Bangalore, India. 1993;May 3:24-28.
- Dellaporta S.L, Wood J, Hicks J.B. A plant DNA minipreparation version II. Plant Mol. Biol. Rep. 1983;1:19-21.
CrossRef - Gradziel T.M. Traditional genetics and breeding. In: Genetics, genomics and breeding of stone fruits. Eds. Kole C and Abbott A.G. CRC Press. 2012;393.
CrossRef - Hartmann H.T, Kester D.E, Davies F.T, Geneve R.L. Hartmann and Kester’s Plant propagation. In: Principles and practices. 5th edition. Prentice-Hall, Englewood Cliffs, N.J. 2002;411-461.
- Janick J. Origins of fruit culture and plant breeding. In: Plant Breeding: The Arnel R. Hallauer International Symposium. Lamkey K.R, Lee M. (Eds.) Blackwell publ, Victoria, Australia. 2006;269-282.
- Liu K, Muse S. Power Marker: new genetic data analysis software, version 2.7 (http://www.powermarker. net); 2004.
- Liu Q, Zhu A, Chai L, Zhou W, Yu K, Ding J, Xu J, Deng X. Transcriptome analysis of a spontaneous mutant in sweet orange [Citrus sinensis (L.) Osbeck] during fruit development. J. Exp. Bot. 2009;60(3):801-813.
CrossRef - Manchekar M.D, Mokashi A.N, Hegde R.V, Venugopal C.K and Byadgi A.S. Clonal variability studies in alphonso mango (Mangifera indica L.) by genetic divergence (D2) analysis. Kar. J. Agric. Sci. 2011;24(4): 490-492.
- Mannin I.F, Gerbi V and Credi R. Heat treated virus-infected grapevine clones: Agronomical and enological modifications. Acta Horti. 1997;473:155-164.
- Mathew P.A and Dhandar D.G. Cardozo Mankurad a breakthrough in mango (Mangifera indica L.) selection. Acta Horti. 1997;455:236-240.
CrossRef - McKey D, Elias M, Pujol B, Duputie, A. The evolutionary ecology of clonally propagated domesticated plants. New Phytol. 2010;186:318–332.
CrossRef - Mukherjee S.K. Systematic and ecogeographic studies of crop gene pools (Mangifera indica L.). Bulletin by International Board for Plant Genetic Resources Rome. 1985;92:235- 240.
- Naik K.C. Improvement of the mango (Mangifera indica L.) by selection and hybridization. Ind. J. Agric. Sci. 1948;18:35-41.
- Negi S.S. Dashehari-51, A regular bearing, high yielding mango clone, ICAR News. 1997;3(4):14.
- Negi S.S. Mango production in India. Acta Horti. 2000;509:9-77.
CrossRef - Oppenheimer C. Study tour report on subtropical fruit growing and research in India and Ceylon. State of Israel. Min. Agric. Rep. Oct. Bulletin. 1956;3.
- Orive M.E. Somatic mutations in organisms with complex life histories. Theor. Pop. Biol. 2001;59:235–249.
CrossRef - Pancaldi M, Weeden N.F, Sansavini S, Mulcahy D.L. Molecular analysis of mutant clones in apple. Acta Horti. 1998;484:311-318.
CrossRef - Patil P.V, Patil V.K, Navale P.A. Grafting in mango. Sci. Horti. 2006;10:45-66.
- Perrier X, Flori A. Bonnot F. Data analysis methods. In: Genetic diversity of cultivated tropical plants. Hamon P, Seguin M, Perrier X, Glaszmann J.C. (Eds.) Enfield, Science Publishers, Montpellier. 2003;43–76.
- Prakash O, Khan R.M. A tryst with mango (Retrospects, Aspects, Prospects). APH Publishing Corporation. 2005;348.
- Prasanth J.M, Reddy P.N, Patil S.R, Pampanagouda B. Effect of cultivars and time of softwood grafting on graft success and survival in mango. Agric. Sci. Digest. 2007;27(1):18-21.
- Ravishankar K.V, Mani B.H, Anand L and Dinesh M.R. Development of new microsatellite markers from Mango (Mangifera indica) and cross-species amplification. Amer. J. Bot. 2011;98:96–99.
CrossRef - Regner F, Wiedeck E, Stadlbauer A. Differentiation and identification of White Riesling clones by genetic markers. Vitis. 2000;42:103-107.
- Singh H, Chadha K.L. Evaluation of some mango cultivars on the basis of their bio-chemical composition. Ind. J. Horti. 1981;38(1-4):70-73.
- Singh R.N, Gorakh S, Rao O.P, Mishra J.S.C. Improvement of ‘Banarasi’, ‘Langra’ through clonal selection. Prog. Horti. 1985;17:273-277.
- Singh S, Gaikwad A.B, Karihaloo J.L. Morphological and molecular analysis of intracultivar variation in indian mango (Mangifera indica l.) Cultivars. Acta Horti. 2009;829:205-212.
CrossRef - Vignani R, Bowers J.E, Meredith C.P. Microsatellite DNA polymorphism analysis of clones of Vitis vinifera, Sangioverse. Sci. Horti. 1996;65:163-169.
CrossRef - Whiley A.W, Mayers P.E, Saranah J, Bartley R.P. Breeding mangoes for Australian conditions. Acta Horti. 1993;341:136-145.
CrossRef - Whitham T.G, Slobodchikoff C.N. Evolution by individuals, plant–herbivore interactions, and mosaics of genetic variability: the adaptive significance of somatic mutations in plants. Oecologia. 1981;49:287–292.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





