How to Cite | Publication History | PlumX Article Matrix
Maryam Dhary Kamel1, Abbas Abdullah Mohammed1 and Ali Abdulhafidh Ibrahim2
1Department of Applied Science, University of Technology, Baghdad, Iraq.
2Department of Business Economics, Al-Nahrain University, Baghdad, Iraq.
Corresponding Author E-mail: maryamdhary1992@yahoo.om
DOI : http://dx.doi.org/10.13005/bbra/2620
ABSTRACT: In the present study, bioinformatics approach has been adopted to explore the sequences and structures analysis of CRP of lung and breast cancer and compares with normal sequence from NCBI. The present study was aimed to investigate the possibility of using CRP as a marker for patients with lung and breast cancer. Also the effect of mutation on the physicochemical properties and structure of CRP. 40 blood and serum samples were examined from patients with lung and breast cancer (aged between 23and 45 years old). Qualitative test was done to detect the presence of CRP in the patient’s serum. The qualitative test showed that 3 (7.5%) patients give positive result and 37(92.5%) patients with lung and breast cancer give negative result to presence of CRP in the serum.In patient with lung cancer five missense mutations and four deletion mutations detected. While in the patient with lung cancer four missense mutations ,six deletion mutations and eight insertion mutation detected by BLAST.One point mutation appeared in all patients at same site and has same effect, this meaning there are relationship between this mutation and cancer disease. This mutation recorded in NCBI, DDBJ and ENA with the numbers LC276938 and LC276937. The present study determined the physico-chemical properties of CRP such as their hydrophilic nature; alpha–helical structure and 3D structure. The results of present study Show that CRP consider non-specific marker for patient with Lung and breast cancer.Also the mutations on CRP gene effected on the structure and physico-chemical properties of C-reactive protein.
KEYWORDS: Bioinformatics Physicochemical; C-reactive Hydrophilic;
Download this article as:| Copy the following to cite this article: Kamel M. D, Mohammed A. A, Ibrahim A. A. Sequence and Structure Analysis of CRP of Lung and Breast Cancer using Bioinformatics Tools and Techniques. Biosci Biotech Res Asia 2018;15(1). |
| Copy the following to cite this URL: Kamel M. D, Mohammed A. A, Ibrahim A. A. Sequence and Structure Analysis of CRP of Lung and Breast Cancer using Bioinformatics Tools and Techniques. Biosci Biotech Res Asia 2018;15(1). Available from: https://www.biotech-asia.org/?p=29229 |
Introduction
C-reactive protein was one of the most important proteins in the medical field and in identifying disease states associated with inflammation. C-reactive protein (CRP), is the one of the most important protein , belonging to pentraxin family of proteins.1 The Cytogenetic location for CRP gene : 1q23.2 which is the long (q) arm of human chromosome1 at position 23.2 ,this position contains gene which encode to CRP2 C-reactive protein produced in the liver in response to IL-6.3 Interlukine-6 consider one of tumour necrosis factor.4 CRP has been associated with cancer size and the stage of disease, and also can it be used as a “cancer marker” in the determining of disease activity.5 Chronic inflammation may be a caused factor in a variety of cancers. In general, the exposure to inflammation for long time, lead to the risk of cancer. The relationship between inflammation and cancer was that central role in the regulation of inflammatory and immune response was IL-6. And also IL-6 plays important roles in cancer progression related to proliferation, migration, and angiogenesis.6 The national cancer institute consider CRP as marker for cancer disease.7 Cancer considers one of the most important health problems of the current era and also a leading cause of death among population. Cancer can simply be defined as a malignant tumor or malignant neoplasm,It includes a group of diseases involving abnormal cell growth with the potential to invade or spread to other parts of the body. It can also be defined as a group of disorders that are characterized by uncontrolled division of cells and the ability of these abnormal cells to spread.8 The most frequent cancer among women was the breast cancer. It’s the female malignant neoplasia with the highest incidence in the world.9 Breast cancer remains a major problem of women’s health and its incidence is increasing in developing countries.10 Another important type of the cancer was the lung cancer. The lung cancer considered the most common cancer in terms of both incidence and mortality .Also it consider the second most commonly occurring of cancer in the world and it is leading to the cancer-related cause of death.11 The aims of this research paper is to investigate the possibility of using CRP as a marker for cancer, computational analysis of CRP using bioinformatics methods, sequence and structural analysis of CRP of breast and lung cancer using bioinformatics methods.
Materials and Methods
Blood Sample Collection
The sample size of this research paper is 40 human patients with breast and lung cancer. For each patient drawn three milliliter of whole blood for patients was obtained under aseptic conditions from each subject by a vein puncture using a disposable syringe. Whole blood was divided into two parts: first part (about two milliliters) was collected in sterile EDTA tube for genetic test. The second part of blood collected in gel tube were separated by centrifugation at 3000 rpm for 10 minutes for qualitative test. The blood and serum samples were subjected to freezing at -20ºC.12
Qualitative Test
The serum samples containing human CRP were obtained from patients with lung and breast cancer, that give positive and negative results for qualitative test by using protocol in the Latex kit for C-Reactive protein (Cat. No.: NS 514001/ Salucea company).
DNA Extraction from Blood Samples
The human DNA extraction from the whole blood of the human patients by using protocol in the gSYNC™ DNA Extraction Kit (Cat. No.: GS100/Genaid company).
Estimation of DNA Concentration and Purity
The human DNA concentration and purity of the samples extracted from whole blood were estimated by using nanodrop(Acta gen/USA) by putting one microliter of the extracted DNA was add to the nanodrop machine to evaluate the concentration in ng/ μl. The purity (1.6-1.8) means DNA high purity.13
Agarose Gel Electrophoresis
After genomic DNA extraction from blood, agarose gel electrophoresis was adopted to confirm the presence and integrity of extracted DNA.13 The samples were carefully loaded into the individual wells of the gel. Then the electrical power was turned on at 56 volt for 30 minutes. Afterwards, the DNA moved from the cathode(-) to the anode(+) poles. The Ethidium Bromaide stained bands in the gel were visualized using ,UV transiluminator at 365 nm and photographed.
Primer Design for CRP gene
The full CRP gene sequences and annotation taken from Genome Database of the national center for biotechnology information (NCBI),the NCBI reference sequence:NC_000001.11.Using primer3 software(http://bioinfo.ul.ee/primer3) for the three Exon of CRP design,14,15 table (1) shows the sequences of the designed for the forward and reverse primers.Then the primers provided by Bioneer Company /Korea in a lyophilized form and using the primers in PCR.
Table 1: Names and sequences of the primers used for CRP gene amplification
| No. | Primer name | Type | Sequence | Size product |
| 1 | CRP1 | Forward | 5′-ATC CAG GCA GGA GGA GGT AG-3′ | 238pb |
| Reverse | 5′-CCC CCA TAC CTC AGA TCA AA-3′ | |||
| 2 | CRP2 | Forward | 5′-TCG AGG AAG GCT TTT GTG TT-3′ | 597pb |
| Reverse | 5′-CTG GGG TTT GGT GAA CAC TT-3′ | |||
| 3 | CRP3 | Forward | 5′-TGT TTG CTT GCA GTG CTT TC-3′ | 485pb |
| Reverse | 5′-TGG GCA GTC CAG GTG TAG AT-3′ |
PCR
The reaction mix was constituted by 30µM KCl,10µM Tris-HCl(PH 9.0),1.5µM MgCl2,250µM each dNTP, 2µl each primer.1U of Taq DNA polymerase , 5 μL of the DNA template(sampl) and ultrapure deionized sterile distilled water up to 20 μL. The thermocycler conditions consisted of the program was set in the thermo cycler to amplify the target DNA for CRP1 ,CRP2 and CRP3 primers in table(2):
Table 2: The program was used for amplification the target DNA for (CRP1 ,CRP2 and CRP3 primers).
| No. | Type of steps | Temperature | Time period | No. of cycles |
| 1 | Pre-denaturation | 94ºC(1) | 5 minutes | 1 |
| 2 | Denaturation | 94ºC(1) | 30 second | 30 |
| 3 | Annealing | 58ºC(2),57ºC(3) | 45 second | |
| 4 | Extension | 72ºC(1) | 1 minute | |
| 5 | Final Extension | 72ºC(1) | 5 minutes | 1 |
| 6 | Storage | 4ºC(1) | ∞ |
Constant for the three primers,
The annealing step for CRP1 and CRP2 primers.
The annealing step for CRP3 primer
Electrophoresis (1 h at 75 V) was performed with 5 μL of the reaction solution in a 2% agarose gel with ethidium bromide. PCR products were visualized under UV light and photographed. The DNA ladder (100pb) was used to estimate the molecular size of the bands.
DNA Sequencing
The result of PCR(Polymerase Chain Reaction) for patient with breast and lung cancer whom gave clear band after PCR analysis of the analyzed (CRP) gene, sent to macrogen company/Korea, for sequencing the three Exon of CRP gene for each samples.
Compared between the DNA sequence of CRP gene for samples and CRP gene retrieved from NCBI
Using BLAST server (https ://blast .ncbi. nlm. nih. G ov / Blast.cgi),16 for detection the location of mutation in CRP for patients.
Translation the DNA sequence of CRP gene to amino acids sequence
Using BLASTX server (https ://blast.ncbi.nlm.nih.Gov/Blast.cgi) for translation DNA nucleotide to amino acids.16
Physicochemical Properties Prediction
Using online tool Prot Param (http://us.expasy.org/tools/protparam.html) for Physico-chemical properties of the CRP for the pateints with lung and breast cancer,also the CRP retraived from NCBI. The parameters computed by Prot Param include the molecular weight, theoretical pI, amino acid composition, atomic composition, instability index, aliphatic index and grand average of hydropathicity (GRAVY).17
Protein Structure Prediction
Using PSIpred online tool (http://bioinf.cs.ucl.ac.uk/psipred/psiform. html) for Secondary structure has been predicting18,19 where the FASTA format of the sequence was given as input. It provides the structural information of the protein sequence in form of coils, helices and using PHYRE2 sofware (http ://www .sbg. bio.ic. ac. uk/ phyre2/ html) for 3D-structure prediction.20 The modeling involves four basic steps, first searching structure showing homology with target, then selecting a best template having maximum identity with the target sequence which follows its alignment with the target and modeling the structure. On the other hand using swiss-model (http://swissmodel.expasy.org/) an automated system for modeling the quaternary structure of a protein from its amino acid sequence using homology modeling techniques21
Results and Discussions
Serum specimens were tested for presence of CRP using latex kit for CRP. However, the results obtained from qualitative test were 1(3.3%) patient give positive result and 29 (96.7%) patients with breast cancer give negative result to CRP, while the study of,22 Which proved that the CRP was act as marker for predictor and to identify the risk of breast cancer. About 2 (20%) patients with lung cancer give positive result and 8(80%) patients give negative result for CRP, while study of,23 that proposed the levels of CRP in serum sample are increased in patients with lung cancer. All these results agree with study of,24 that proved CRP was non-specific marker for inflammation. Genomic DNA from the blood samples was extracted following the standard protocol which used in many researches and genetic studies according to.13 The quantification of DNA measured by Nanodrop had revealed that the DNA concentration ranged(3-5) ng/μl and purity ranged was (1.6-1.8).The result demonstrated that the purity of the extracted DNA in all samples were sufficiently high for PCR analysis.
PCR
The present study used PCR technique for DNA isolated from the blood from patients to amplify the 3 exons of the CRP gene.By using three primers(CRP1,CRP2 and CRP3 primer) ,that were designe by primer3 software and obtained from Bioneer Company (Korea). The PCR results were interpreted by the presence or absence of specific bands of amplified gene on 2% agarose. In this study, three primers were screened for PCR analysis using 40 DNA samples that were extraction from the whole blood of patients. The results of using first set of primers (CRP1F and CRP1R), second set of primers (CRP2F and CRP2R) and using third set of primers (CRP3F and CRP3RR) showed amplified fragment (238pb,597bp and485bp. respectively) a clear band by electrophoresis on a 2% agarose gel at 60 volt for 90 minute, of 40 DNA samples as shown in figure (1).
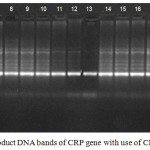 |
Figure 1: Detection of PCR product DNA bands of CRP gene with use of CRP1F CRP1R primers (238 bp.).
|
The amplified fragment were separated by electrophoresis on a 2% agarose gel, stained with ethidium bromide at 60 volt for 90 minute. Photographed under UV light.
Lanes (1-10) patient with breast cancer gave band for amplified fragment.
Lanes (11-20) patients with lung cancer gave band for amplified fragment.
M:Marker DNA ladder size (100bp)
The result of using second primer (CRP2F and CRP2R) showed amplified fragment (597bp.) as a clear band by electrophoresis on a 2% agarose gel at 60 volt for 90 minute, as shown in figure (2) in all patients and control.
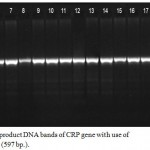 |
Figure 2: Detection of PCR product DNA bands of CRP gene with use of CRP2F and CRP2R primers (597 bp.).
|
The amplified fragment were separated by electrophoresis on a 2% agarose gel, stained with ethidium bromide at 60 volt for 90 minute. Photographed under UV light.
Lanes (1-10) patient with breast cancer gave band for amplified fragment.
Lanes (11-20) patients with lung cancer gave band for amplified fragment.
M: Marker DNA ladder size (100bp)
The result of using third set of primer (CRP3F and CRP3RR) showed amplified fragment (485bp.) as a clear band by electrophoresis on a 2% agarose gel at 60 volt for 90 minute, as shown in figure (3) in all patients and control.
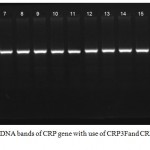 |
Figure 3: Detection of PCR product DNA bands of CRP gene with use of CRP3Fand CRP3R primers (485 bp.).
|
The amplified fragment were separated by electrophoresis on a 2% agarose gel, stained with ethidium bromide at 60 volt for 90 minute. Photographed under UV light.
Lanes (1-10) patient with breast cancer gave band for amplified fragment.
Lanes (11-20) patients with lung cancer gave band for amplified fragment.
M: Marker DNA ladder size (100bp)
The outcomes of the CRP gene amplification using PCR analysis represented that all patients gave positive result (formation bands). This results showed that the product of amplified first set of primers (CRP1F and CRP1R), second set of primers (CRP2F and CRP2R) and third set of primers (CRP3F and CRP3RR) of CRP gene find in all patients.
Sequencing of C-reactive protein gene
The current study utilized forward primers for two patients by direct sequencing: first with lung cancer4 and second patient with breast cancer figure(5).
 |
Figure 4: The chromatogram for the forward nucleotide sequence of amplified the first primer for patient with lung cancer by DNA sequencer.
|
 |
Figure 5: The chromatogram for the forward nucleotide sequence of amplified the first primer for patient with breast cancer by DNA sequencer.
|
The comparison between DNA subjects and reference sequence are shown in figure(6).
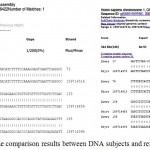 |
Figure 6: shown simple part of the comparison results between DNA subjects and reference sequence by BLAST.
|
Genotype analysis of CRP gene for the PCR product of the primer indicated of much genetic alteration. The effect of mutations in translation and the result revealed that there were five missense mutations and four deletion mutations in patient with breast cancer. The foure missense mutations, six deletion mutations and eight insertion mutation in the patient with lung cancer table.3
Table 3: Types of mutations for the Exon of CRP gene sequencing.
| Types of disease | No. of primer | Wild genetic code of DNA | Mutations in genetic code of DNA | Type of mutations at DNA level | Wild genetic code of RNA | Mutations in genetic code of RNA | Effect of translation | Site of mutations |
| Breast cancer | First | GAA | GTA | Transversion | GAA Glutamate | GUA Valine | Missense mutation | 159714558 |
| Breast cancer | First | GCC | G – C | Deletion mutation | GCC Alanine | G – C | Frame shift Mutation | 159714564 |
| Breast cancer | First | AGA | ACA | Transversion | AGA Arginine | ACA Threonine | Missense mutation | 159714397 |
| Breast cancer | First | ATG | AAG | Transversion | AUG Methionine (start code) |
AAG Lysine | Missense mutation | 159714441 |
| Breast cancer | Second | AAG | A – G | Deletion mutation | AAG Lysine | A – G | Frame shift mutation | 159714077 |
| Breast cancer | Second | CCA | CAA | Transversion | CCA Proline | CAA Glutamine | Missense mutation | 159713546 |
| Breast cancer | Third | AAA | A – – | Deletion mutation | AAA Lysine | A – – | Frame shift mutation | 159712764 (-) 159712763(-) |
| Breast cancer | Third | TGT | TAT | Transition | UGU Cysteine | UAU Tyrosine | Missense mutation | 159712443 |
| Breast cancer | Third | CTG | C – G | Deletion mutation | CUG Leucine | C – G | Frame shift mutation | 159712333 |
| Lung cancer | First | GCC | G-C | Deletion mutation | GCC Alanine | G-C | Frame shift mutation | 159714564 |
| Lung cancer | First | GAA | GTA | Transversion | GAA Glutamate | GUA Valine | Missense mutant | 159714558 |
| Lung cancer | First | AGA | ACA | Transversion | AGA Arginine | ACA Threonine | Missense mutant | 159714397 |
| Lung cancer | First | ATG | AAG | Transversion | AUG Methionin (start code) |
AAG Lysine | Missense mutant | 159714441 |
| Lung cancer | First | GAG | GGG | Transition | GAG Glutamate | GGG Glycine | Missense mutant | 159714386 |
| Lung cancer | Second | TCA | T– | Deletion mutations | UCA Serine | U– | Frame shift mutation | 159714056 159714055 |
| Lung cancer | Second | CTC | C-A | Deletion and transvertion mutations | CUC Leucine |
C-A | Frame shift mutation | 159714013(-) 159714012(A) |
| Lung cancer | Second | C—TT | CTTT | Insertion mutations | C–UU Leucine | CUUUU Leucine | Frame shift mutation | Between two site 159713619 159713620 |
| Lung cancer | Second | G-TC | GGT | Insertion mutation | G-UC Valine |
GGUC Glycine |
Frame shift mutation | Between two site 159713595 159713596 |
| Lung cancer | Second | A-TC | AAT | Insertion mutation | A-UC Isoleucine |
AAUC Asparagines |
Frame shift mutation | Between two site 159713626 159713627 |
| Lung cancer | Second | G-CG | GGC | Insertion mutations | G-CG Alanine | GGCG Glycine | Frame shift mutation | Between two site 159713558 159713559 |
| Lung cancer | Second | T—GA | TTGG | Insertion mutations | U—GA Stop codon |
UUGGA Leucine |
Frame shift mutation | Between two site 159713576 159713577 |
| Lung cancer | Second | G—CG | GGGC | Insertion mutations | G—CG Alanine | GGGCG Glycine | Frame shift mutation | Between two site 159713584 159713585 |
| Lung cancer | Second | T-CC | TTC | Insertion mutation | U-CC Serine | UUCC Phenylalanine | Frame shift mutation | Between two site 159713594 159713595 |
| Lung cancer | Second | G-TC | GGT | Insertion mutation | G-UC Valine | GGUC Glycine | Frame shift mutation | Between two location 159713603 159713604 |
| Lung cancer | Second | CAC | C-C | Deletion mutation | CAC Histidine | C-C | Frame shift mutation | 159713549 |
| Lung cancer | Third | CTT | C-T | Deletion mutation | CUU Leucine | C-U | Frame shift mutation | 159712772 |
| Lung cancer | Third | GAA | G– | Deletion mutations | GAA Glutamate | G– | Frame shift mutation | 159712765 159712764 |
Mutation
Genetic code for amino acid produces from insertion mutation
Finally the thymine transversion by adenine at the site 159714441 in the first Exon of CRP gene on the long arm of first chromosome. This mutation recorded in NCBI ,DDBJ and ENA with the numbers LC276937 and LC276938 this point mutation effected on the translation and cause missense mutation , when methionine replaced by lysine appeared in all patients at same site and has same effect, this meaning there are relationship between this mutation and cancer disease. And also this mutation detection in multiple tumor samples, so it can be consider the cause of cancer disease these result agree with study25 demonstrated that Tay–Sachs disease is caused by a genetic mutation in the HEXA gene on chromosome 15. these result agree with study26 demonstrated that sickle-cell anemia is caused by a point mutation in the β-globin chain of hemoglobin, caused replace glutamic acid with amino acid valine at the sixth position. these result agree with studies of27,28 demonstrated that neurofibromatosis is caused by point mutations in the neurofibromin gene and these result agree with study29 demonstrated that cystic fibrosis is caused by a mutation in the CFTR gene.
Primary structure of C-reactive protein
The sequence of CRP for patients with lung and breast cancer consist from 219 and 211 amino acids respectively. While the sequence of CRP retrieved from NCBI consist from 224 amino acids.
Sequence Analysis
Primary structure analysis provided the physicochemical properties of CRP.Molecular weight of CRP for the patients with lung and breast cancer was (24338.88 and 23508.82 MW). While the molecular weight for CRP that retrieved from NCBI FASTA format-protein was (25038.58MW). Most abundant amino acid was Ser(S) ) in CRP for lung and breast cancer 9.6% and 9.5% respectively, while in CRP that retrieved from NCBI was 10.3%, Gly(G) 8.7% and 8.5% respectively, while in CRP that retrieved from NCBI was 8.0%, Leu(L) 8.7% and 8.5% respectively, while in CRP that retrieved from NCBI was 8.9%, val (V) 8.2% and 8.5% respectively, while in CRP that retrieved from NCBI was 8.5%. A protein whose instability index is smaller than 40 is predicted as stable, a value above 40 predicts that the protein may be unstable.20 Prot Param server predicted that CRP for patients with lung and breast cancer were stable. And also the CRP that retrieved from NCBI was stable. The isoelectric point of a protein is an important property, because it is at this point that the protein is least soluble.20 Computed isoelectric point of CRP for patients with lung and breast cancer were 5.75 and 5.26 respectively also the PI for CRP that retrieved from NCBI was 5.76 below 7 so they are likely to precipitate in acidic buffers. This result agree with studies of30,31 demonstrated that the mutation cause large changes in the sequences this effected on physiochemical properties of protein especially on protein stability.
Structure Analysis
The secondary structure of present study gave the following alpha helix predicted values 6.06%,6.06% ,5.71% and 6.06% for CRP of patients with lung, breast cancer and CRP that retrieved from NCBI respectively. Secondary structure of present study gave the following ß- turn predicted values 42.42%, 42.86% and 42.42% for CRP of patients with lung ,breast cancer and CRP that retrieved from NCBI respectively. Secondary structure of present study gave the following coil predicted values 52.5% , 51.52% and51.52% for CRP of patients with lung ,breast cancer and CRP that retrieved from NCBI respectively figure.7
 |
Figure 7: Secondary structure of CRP for patient with lung cancer(A) ,breast cancer(B) and CRP that retrieved from NCBI(C) respectively by PSIpred.
|
For CRP of patient with lung cancer, out of all the templates given by PHYRE2 server, the one with highest % i.d. (94 %) was used to predict the 3D structure. For CRP of patient with breast cancer, out of all the templates given by PHYRE2 server, the one with highest % i.d. (99 %) was used to predict the 3D structure. For CRP retrieved from NCBI, out of all the templates given by PHYRE2 server, the one with highest % i.d. (100 %) was used to predict the 3D structure figure.8
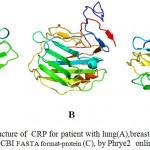 |
Figure 8: 3D structure of CRP for patient with lung(A),breast cancer(B) and CRP retrieved from NCBI FASTA format-protein (C), by Phrye2 online software.
|
The N-termini of proteins are colored blue and the C –termini red.
For CRP of patient with lung cancer, out of all the templates given by swiss-model server, the one with highest % i.d.(94.53 %) was used to predict the quaternary structure . For CRP of patient with breast cancer, out of all the templates given by swiss-modle server, the one with highest % i.d.(98.96 %) was used to predict the quaternary structure. For CRP retrieved from NCBI, out of all the templates given by swiss-modle server, the one with highest % i.d. (100 %) was used to predict the quaternary structure figure(9).
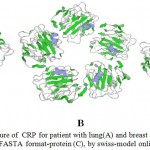 |
Figure 9: Quaternary structure of CRP for patient with lung(A) and breast cancer(B) and CRP retrieved from NCBI FASTA format-protein (C), by swiss-model online software.
|
The α-helix of protein is colored purple, β-sheets colored green and the coil light gray.
The mutations on CRP gene for patients with lung and breast cancer were recorded by present study caused different effects on the structure of protein when it compared with CRP retrieved from NCBI as show in the result of structure analysis such as the numbers and positions of alpha helix, ß- turn and coil as shown in figures(7,8 and 4), this result agree with studies30,31 demonstrated that mutation cause large changes in the sequences this effected on the structurally of protein. Furthermore, this study has been shown that pathways of protein folding are largely unaffected by changes in the sequence.
Conclusions
In the present study, CRP is non-specific marker for patients with breast and breast cancer. The genotype at the site 159714441 was found to be associated with cancer disease. The mutations on CRP gene for patients with lung and breast cancer effect on the physicochemical properties of C-reactive protein such as molecular weight, PI, percentage of amino acid and also protein stability when compared with CRP retrieved from NCBI. The mutation effected on structures of CRP for the present study by changed the number and position of alpha helix, ß- turn and coil this effected on CRP by decrease 2Ca+2 ion for each subunit ,this lead to loss the function of host defense of CRP for patients with lung and breast cancer.
References
- Srikantiah C. C – reactive protein: An inflammatory marker with specific role in physiology, pathology, and diagnosis. Internet Journal of Rheumatology and Clinical Immunology. 2014;2(1):1-23.
- Pepys M and Hirschfield G. C-reactive protein: a critical update. Journal of Clinical Investigation. 2003;111(12):1805-1812.
CrossRef - SrikantiahC. C – reactive protein: An inflammatory marker with specific role in physiology, pathology, and diagnosis. Internet Journal of Rheumatology and Clinical Immunology. 2014;2(1):1-23.
- Mazhar D and Ngan S. C-reactive protein and colorectal cancer. Quarterly journal of medicine. 2006;99(8):555–559.
CrossRef - Davis V and Rose P. Adipokines as endocrine, paracrine, and autocrine factors in breast cancer risk and progression. Endocrine-related Cancer. 2007;14:189–206.
CrossRef - Nagasaki T, Hara M, Shiga K and Takeyama H. Relationship between inflammation and cancer progression: review of the recent advances in interleukin-6 signaling and its blockage in cancer therapy. Receptors & Clinical Investigation. 2014;1(3):1-7.
- Kao P, Shu-Chu S and Ta-Jen W.u . Serum C-Reactive Protein as a Marker for Wellness Assessment . Annals of Clinical & Laboratory Scienc . 2006;36(2):163-169.
- Blachford S.L. The Gale Encyclopedia of Genetic Disorders. Detroit Gale Group. Thomsonlearning.U.S.A. 2002;1:1345.
- Maccio A, Madeddue C and Mantovani G. Adipose tissue as target organ in the treatment of hormone-dependent breast cancer:new therapeutic perspectives. International Association for the Study of Obesity. 2009;10;660-670.
CrossRef - Guo L, Liu S, Zhang S, Chen Q, Zhang M, Quan P, Lu J and Sun X. C-reactive protein and risk of breast cancer: A systematic review and meta-analysis. International Journal of Scientific Reports. 2015;5(10508):1-8.
- Thanveer V and Balaji M. Protein Modeling and Sequence Analysis Studies on Human Lung Cancer Potential Protein Target (Crp C-Reactive Protein,Pentraxin-Related)Using Bioinformatics Protocols. International Journal of Pharmaceutics. 2012;2(4):166-173.
- Qiling L, Kang T, Tian X,Yamin A, Min L, Richard J, Bythwood T, Wang Y, Li X, Liu D, Ma L and Song Q. Multimeric Stability of Human C-reactive Protein in Archived Specimens. Public Library of Science. 2013;8(3):1-6.
- Sambrook J and Russel D. Molecular cloning: a laboratory manual.3ed ed.Cold Spring Harbor,New York: Cold Spring Harbor laboratory press. 2001.
- Aggarwal P. Bistro-Primer – Tool to design and validate specific PCR primer pairs for phylogenetic analysis. Marquette University. 2009;1-79.
- Jian X, Coulouris G, Zaretskaya I,Cutcutache I, Rozens S and Madden T. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BioMed Central. 2012;13(134):1-11.
- Agostino M. Practical Bioinformatics.Garland Science. 2013;47-60.
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins M, Appel R, Bairoch A and Walker J. The Proteomics Protocols Handbook. Humana Press. 2005;571-607.
CrossRef - Buchan A.W, Minneci F, Nugent O.C, Bryson K and Jones T. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Research. 2013;41:349-357
CrossRef - Davies A, Rigden J, Phelan M and Madine J. Probing Medin Monomer Structure and its Amyloid Nucleation Using 13C-Direct Detection NMR in Combination with Structural Bioinformatics. Scientific Reports. 2017;7( 45224):1-10.
CrossRef - Jain D, Rawat R and Jatav K.V. Phylogenetic analysis of RAS subfamily proteins . International Journal of Pharma Sciences and Research. 2016;7(3):1070-1080.
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino T, Bertoni M, Bordoli L and Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research. 2014;42(1):252-258.
CrossRef - Asegaonkar B, Asegaonkar N, Takalkar V, Advani S and Thorat P. C-Reactive Protein and Breast Cancer:New Insights from Old Molecule. International Journal of Breast Cancer.201;2015(145647):1-7.
- Aref H and Refaat S. CRP evaluation in non-small cell lung cancer. Egyptian Journal of Chest Diseases and Tuberculosis. 2014;63:717-722.
CrossRef - Mazha D and Ngan S. C-reactive protein and colorectal cancer. Quarterly Journal of Medicine. 2006;99:555–559.
CrossRef - Myerowitz R. Tay-Sachs disease-causing mutations and neutral polymorphisms in the Hex A gene. Human Mutation . 1997;9(3):159-208.
CrossRef - Hisa C. Respiratory function of hemoglobin. The New England Journal of Medicine. 1998;338(4):239-247.
- Serra E, Ars E, Ravella A, Sanchez A, Puig S, Rosenbaum T, Estivill X and Lazaro C. Somatic NF1 mutational spectrum in benign neurofibromas: mRNA splice defects are common among point mutations. Springer link. 2001;108(5):416-429.
- Wiest V,Eisenbarth I,Schmegner C,Krone W and Assum G. Somatic NF1 mutation spectra in a family with neurofibromatosis type 1: Toward a theory of genetic modifiers. Human Mutation. 2003;22(6):423-427.
CrossRef - Bobadilla L,Macek M,Fine P and Farrell M. Cystic fibrosis: A worldwide analysis of CFTR mutations-Correlation with incidence data and application to screening. Human Mutation. 2002;19(6):575-606.
CrossRef - Nussinov R and Sinha N. Point mutations and sequence variability in proteins:Redistributions of preexisting populations. Proceedings of the National Academy of Sciences of the United States of America. 2000;6(98):3139-3144.
- Venselaar H, Beek T, Kuipers K, Hekkelman L and Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BioMed Central. 2010;11(548):1-10.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





