How to Cite | Publication History | PlumX Article Matrix
Genetic Susceptibility to Breast cancer in East Azerbaijan, Iran
Mahdiyeh Pashaei1,3, Jamal Eivazi Ziaei1, Alireza Nikanfar1, Babak Emamalizadeh2 and Seyyed Mojtaba Mohaddes Ardebili1,3
1Hematology and Oncology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
2Department of Medical Genetics, Tabriz University of Medical Sciences, Tabriz, Iran.
3Department of Biochemistry and Clinical Laboratories, Division of Medical Genetics, Tabriz University of Medical Sciences, Tabriz, Iran.
Corresponding Author E-mail: mojtabamohaddesardebili197@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2651
ABSTRACT: Breast cancer is the most common cause of death among women in the world and in Iran. A number of risk factors for breast cancer development have been identified, among which the most important is positive family history. Alterations in different genes, including BRCA1, BRCA2, p53, CHEK2, PTEN, and ATM, also induce a predisposition for breast cancer. Among these changes, BRCA1 and BRCA2 alterations are the strongest drivers of breast cancer predisposition. This study was aimed at contributing to the development of appropriate methods for detecting genetic alterations, such as single or multiple exon deletions and amplifications, in the aforementioned genes. We used multiplex ligation-dependent probe amplification (MLPA) to determine genetic alterations in 150 female patients who hail from East Azerbaijan, Iran and suffer from familial breast cancer. Specifically, we investigated copy number changes in BRCA1, ATM, p53, CHEK2, and PTEN. MLPA results showed no remarkable mutations in the study population. Size coverage is a critical factor for MLPA to accurately detect potential mutations in familial breast cancer susceptibility genes.
KEYWORDS: Breast Cancer; BRCA1; CHEK2; MLPA; PTEN; p53
Download this article as:| Copy the following to cite this article: Pashaei M, Ziaei J. E, Nikanfar A, Emamalizadeh B, Ardebili S. M. M. Genetic Susceptibility to Breast cancer in East Azerbaijan, Iran. Biosci Biotech Res Asia 2018;15(2). |
| Copy the following to cite this URL: Pashaei M, Ziaei J. E, Nikanfar A, Emamalizadeh B, Ardebili S. M. M. Genetic Susceptibility to Breast cancer in East Azerbaijan, Iran. Biosci Biotech Res Asia 2018;15(2). Available from: https://www.biotech-asia.org/?p=30151 |
Introduction
Breast cancer accounts for about one-fourth of female cancer cases worldwide. It is the most frequently occurring malignancy and the second cause of death among Iranian women.1 Different factors, such as gender, ethnic origin, and age-specific patterns, are recognized as important predisposing factors for breast cancer. Compared with sporadic breast cancer, which has an estimated incidence frequency ranging from 90% to 95%, familial breast cancer has been estimated to occur only at 5% to 10% frequency. Nevertheless, the most frequently characterized predisposing factor for the disease is positive family history.2 Interestingly, less than 10% of breast cancer types are attributable to a single highly penetrant inherited predisposing allele.3 Less than a quarter of familial risk factors have been related to mutations in identified breast cancer genes; the rest remain unrecognized. Mutation screening based on candidate gene approaches and genome-wide association studies have led to unique classifications of breast cancer predisposing alleles; these classifications are (a) high-penetrance alleles, (b) rare moderate-penetrance alleles, and (c) common low-penetrance alleles, which have varying prevalence rates in different populations.4 Additional mutations that cause breast cancer susceptibility have remained unidentified. Such mutations may occur in moderate- to low-penetrance gene variants that may relatively increase breast cancer risk for carriers through multiplicative and/or cumulative effects.5 Conversely, each single variant may individually impose minimal risk.
BRCA1 (breast cancer susceptibility gene 1) and BRCA2 (breast cancer susceptibility gene 2) are the most important cancer susceptibility genes. BRCA1 and BRCA2 mutations account for about 20% to 24% of hereditary breast cancer in females6 and BRCA1 is a major causative gene for early-onset breast cancer.7 Mutations in these genes may result in faulty DNA repair that possibly leads to malignancy in cases of high mutation rates.8 Among breast cancer patients with no family history of the disease, 65% are carriers of BRCA1 mutations, whereas the remaining 35% are carriers of BRCA2 mutations.9 Genetic variants of undefined conditions are majorly missense mutations and minorly in-frame deletions.3
The fact that additional high-penetrance susceptibility genes could not be identified via genome-wide linkage studies in non-BRCA1/BRCA2 families shows that BRCA1 and BRCA2 account for a very small fraction of familial breast cancer incidences. Cancer-predisposing syndromes, such as Li-Fraumeni, Peutz-Jegher, Cowden, and Neurofibromatosis disease, present increased risk of breast cancer; studies on these disorders have shown that mutations in p53 (protein 53), STK11 (serine/threonine kinase 11), PTEN (phosphatase and tensin homolog), NF1 (neurofibromatosis type 1) and CDH1 (cadherin-1) can increase breast cancer risk.10,11
Despite various efforts to detect other highly penetrant breast cancer predisposing genes, no study has identified such role for BRCA3 gene. However, the sequencing of genes involved in DNA repair presented opportunities to identify several intermediate-penetrance breast cancer susceptibility genes, such as CHEK2 (checkpoint kinase-2), BRIP1/BACH1, PALB2, PTEN, ATM, and p53, most of which function as cell cycle controllers, DNA integrity insurers, and signal transducers.12,13
Large genomic rearrangements (LGRs) are widely expected in a considerable percentage of breast cancer types in various populations. The LGRs of BRCA1 and BRCA2 have been observed in many cases of breast cancer in which no genetic alterations were identified beforehand using common screening methods.14,15 An LGR within the CHEK2 gene has been reported to occur among Finnish, Northern European, Mayo, French, and American people. The first evidence of a large CHEK2 duplication was found primarily as a predisposing risk factor in an Italian family that showed a hereditary pattern of breast cancer. The family had a 23 kb duplicated region in the CHEK2 gene spanning intron 5 to 13.16 An LGR in the p53 gene has also been observed in the Brazilian population. Common mutations, such as 1100delC, I157T, and IVS2 + 1G > A, were reported in CHEK2 and attributed to a variety of malignancies. The most common types of alterations have been reported as small insertions/deletions leading to an entirely non-functional BRCA protein (Figure 1). The higher rate of duplication/deletion in BRCA1 than in BRCA2 was attributed to the accumulation of Alu (arithmetic logic unit) sequences.17 LGRs that typically result from homologous recombination between BRCA1 and pseudogenes (genes with similar sequences) compose about one-third of all the mutations that take place in the BRCA1 gene.
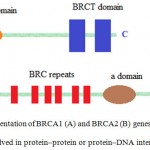 |
Figure 1: Schematic representation of BRCA1 (A) and BRCA2 (B) genes demonstrating functional domains involved in protein–protein or protein–DNA interactions (16).
|
The importance of LGRs in breast cancer development, the failure of routine screening assays to detect such cases, and the presence of numerous undetected breast cancer cases in East Azerbaijan, Iran led us to hypothesize a high prevalence of LGRs in breast cancer cases in this region. Accordingly, we investigated the presence of LGRs in CHEK2, ATM, PTEN, p53, and BRCA1 by using multiplex ligation-dependent probe amplification for a group of East Azerbaijan breast cancer cases.
Materials and Methods
One hundred and fifty available breast cancer cases referred by an oncologist were recruited as a case group. All the cases satisfied international BOADICEA standard criteria for cancer assessment. Sixteen healthy women from the same area were recruited as the control group. Written informed consent (code: 915071) was obtained from all the participants. After sample collection, genomic DNA was extracted using the salting-out method.
MLPA reaction was carried out in four steps: (A) denaturation and hybridization of MLPA probes, (B) ligation, (C) polymerase chain reaction (PCR) amplification, and (D) separation and purification of PCR products. Differences in probe amplifications were identified by comparing the signal peaks of the case and control samples. The comparison of the probes with different amplification signals and the references revealed sequences with aberrant copy numbers. Oligonucleotide probes that were not ligated to the target could not be amplified and, thus, did not produce any signal. Alternatively, the amplification of the genes enabled us to establish additional templates for MLPA probes and generate additional PCR products for comparison with the normal references.
The loss and gain of DNA material within BRCA1, CHEK2, p53, PTEN, and ATM were investigated in the case and control groups by using a P190-C1 kit (MRC-Netherlands). An ABI3100 genetic analyzer was used to purify the products, and GENEMARKER V.2.6.0 was used to analyze the results.
The control probe curves were directly normalized using the median of all the samples, and two standard deviations were used as normalization factors. Normal peak ratio was considered to be 1, and potential duplication and deletion were regarded as peak ratios ≥1.3 and ≤0.7, respectively. Positive controls were used for all the detected modifications. This study was approved by the ethical board committee of Tabriz University of Medical Sciences.
Results
The kit used in this study comes with 43 probes for CHEK2, including 16 probes for exon 1 to 16 and one extra probe for exon 11. However, the kit provides no probe for second and third alternative exons. The number of probes used to investigate alterations in the ATM gene, PTEN gene, flanking KLLN gene, p53 gene, and BRCA1 promoter were seven, five, one, two, and one, respectively. Eight probes were included in the probe mix as reference for the detection of a few altered autosomal chromosome loci. We analyzed 150 samples related to familial breast cancer and high-risk cases. Interestingly, no conclusive new LGR was found in BRCA1, CHEK2, p53, ATM, and PTEN (Figure 2). The results are highly concordant with the findings of an Italian study (28, 34). Note that the P190-C1 kit has only one BRCA1 gene probe designed to check the promoter region, which was not covered the hotspots. This exclusion may be one of the reasons for the absence of mutations in the studied population.
 |
Figure 2: MLPA results for the CHEK2, p53, BRCA1, ATM, and PTEN genes of a breast cancer case.
|
Discussion
A total of 150 familial breast cancer cases from East Azerbaijan, Iran were randomly selected for this research. LGRs in the CHEK2, ATM, PTEN, p53, and BRCA1 genes of the study population were investigated via MLPA. Three families exhibited a 1100delC mutation in the kinase domain of the CHEK2 gene. This mutation results in the early termination of the chain during translation.18 Previous studies indicated that mutations such as 1100delC in the CHEK2 gene are associated with the development of several types of cancer in different populations.19–21 Using MLPA, other studies demonstrated that deletions/duplications in the BRCA1 gene can lead to breast cancer in various populations. On the basis of our experimental results, we propose that LGRs in the CHEK2, p53, ATM, BRCA1, and PTEN genes are unlikely to exert an important effect on the etiology of breast cancer in East Azerbaijan, Iran.22,23
Our findings are similar to the results of studies on CHEK2 1100delC in Korean, Chinese, Japanese, South Indian, and Singaporean populations. The results of the current work are also highly similar to those of a study on BRCA1 and BRCA2 LGRs in breast cancer cases from Sri Lanka. Other studies likewise revealed no p53 mutation in breast cancer cases.17
In summary, LGRs may not be found in selected regions of the CHEK2, ATM, PTEN, p53, and BRCA1 genes. The coverage size of the MLPA kit is a critical factor for similar studies to accurately detect potential mutations in familial breast cancer susceptibility genes. Using a kit with better coverage for breast cancer hotspots can be useful in identifying large deletions and duplications. Additional investigations are required to uncover genomic rearrangements in other regions of breast cancer predisposing genes for the East Azerbaijan context.
References
- Jafari-Koshki T, Mansourian M, Mokarian F. Exploring factors related to metastasis free survival in breast cancer patients using Bayesian cure models. Asian Pac .J. Cancer Prev. 2014;15:9673-9678.
CrossRef - Dumitrescu R.G, Cotarla I. Understanding breast cancer risk – where do we stand in 2005? J. Cell Mol Med. 2005;9:208-221.
CrossRef - Oldenburg R.A, Meijers-Heijboer H, Cornelisse C.J, Devilee P. Genetic susceptibility for breast cancer: how many more genes to be found? Crit Rev Oncol Hematol. 2007;63:125-149.
CrossRef - Berliner J.L, Fay A.M, Group PISotNSoGCFCRCSI. Risk assessment and genetic counseling for hereditary breast and ovarian cancer: recommendations of the National Society of Genetic Counselors. J. Genet Couns 2007;16:241-260.
CrossRef - Easton D.F. How many more breast cancer predisposition genes are there? Breast Cancer Res. 1999;1:14-17.
CrossRef - Petrucelli N, Daly M.B, Feldman G.L. Hereditary breast and ovarian cancer due to mutations in BRCA1 and BRCA2. Genet Med. 2010;12:245-259.
CrossRef - Kuusisto K.M, Bebel A, Vihinen M, Schleutker J, Sallinen S.L. Screening for BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish BRCA1/2-founder mutation-negative breast and/or ovarian cancer individuals. Breast Cancer Res. 2011;13:20.
CrossRef - Prucka S.K, McIlvried D.E, Korf B.R. Cancer risk assessment and the genetic counseling process: using hereditary breast and ovarian cancer as an example. Med Princ Pract. 2008;17:173-189.
CrossRef - Antoniou A, Pharoah P.D, Narod S, Risch H.A, Eyfjord J.E, Hopper J.L, Loman N, Olsson O, Johannsson O, Borg A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am.J. Hum Genet. 2003;72: 1117-1130.
CrossRef - Ray M.E, Yang Z.Q, Albertson D, Kleer C.G, Washburn J.G, Macoska J.A, Ethier S.P. Genomic and expression analysis of the 8p11-12 amplicon in human breast cancer cell lines. Cancer Res. 2004;64:40-47.
CrossRef - Frank S.A. Genetic predisposition to cancer – insights from population genetics. Nat .Rev.Genet. 2004;5:764-772.
CrossRef - Seitz S, Rohde K, Bender E, Nothnagel A, Kölble K, Schlag P.M, Scherneck S. Strong indication for a breast cancer susceptibility gene on chromosome 8p12-p22: linkage analysis in German breast cancer families. Oncogene. 1997;14:741-743.
CrossRef - Wang Y, Dai B, Ye D. CHEK2 mutation and risk of prostate cancer: a systematic review and meta-analysis. Int J Clin Exp Med. 2015;8:15708-15715.
- Yassaee V.R, Emamalizadeh B, Omrani M.D. Screening for genomic rearrangements at BRCA1 locus in Iranian women with breast cancer using multiplex ligation-dependent probe amplification. J. Genet. 2013;92: 131-134.
CrossRef - Ewald I.P, Ribeiro P.L, Palmero E.I, Cossio S.L, Giugliani R, Ashton-Prolla P. Genomic rearrangements in BRCA1 and BRCA2: A literature review. Genet Mol Biol. 2009;32:437-446.
CrossRef - Tedaldi G, Danesi R, Zampiga V, Tebaldi M, Bedei L, Zoli W,Amadori D, Falcini F, Calistri O. First evidence of a large CHEK2 duplication involved in cancer predisposition in an Italian family with hereditary breast cancer. BMC Cancer. 2014;14:478.
CrossRef - Achatz M.I, Hainaut P, Ashton-Prolla P. Highly prevalent TP53 mutation predisposing to many cancers in the Brazilian population: a case for newborn screening? Lancet Oncol. 2009;10:920-925.
CrossRef - Zhang J, Fackenthal J.D, Huo D, Zheng Y, Olopade O.I. Searching for large genomic rearrangements of the BRCA1 gene in a Nigerian population. Breast Cancer Res Treat. 2010;124:573-577.
CrossRef - Walsh T, Casadei S, Coats K.H, Swisher E, Stray S.M, Higgins J, Roch K.C,Madell J,Lee M.K , Ciernikova S. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006;295:1379-1388.
CrossRef - Mazoyer S. Genomic rearrangements in the BRCA1 and BRCA2 genes. Hum Mutat. 2005;25:415-422.
CrossRef - Montagna M, Dalla Palma M, Menin C, Agata S, De Nicolo A, Chieco-Bianchi L, D’Andrea E. Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet. 2003;12:1055-1061.
CrossRef - Mohamad S, Isa N.M, Muhammad R, Emran N.A, Kitan N.M, Kang P, Kang I.N, Mohd Taib N.A, Wang S.H.T, Akmal N, Sh N. Low prevalence of CHEK2 gene mutations in multiethnic cohorts of breast cancer patients in Malaysia .PLoS One. 2015;10:e0117104.
CrossRef - Puget N, Gad S, Perrin-Vidoz L, Sinilnikova O.M, Stoppa-Lyonnet D, Lenoir G.M, Mazoyer S. Distinct BRCA1 rearrangements involving the BRCA1 pseudogene suggest the existence of a recombination hot spot. Am J Hum Genet. 2002;70:858-865.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





