How to Cite | Publication History | PlumX Article Matrix
Gothandapani Sellamuthu1,2, Prabha Shankar1, Sekar Soundarapandian2, Sangeeta Paul3 and Jasdeep C. Padaria1
1ICAR - National Research Centre on Plant Biotechnology, Pusa Campus, New Delhi- 110012, India.
2Department of Industrial Biotechnology, Bharathidasan University, Tiruchirappalli – 620 024, India.
3ICAR – Indian Agricultural Research Institute, Pusa Campus, New Delhi- 110012, India.
Corresponding Author E-mail: jasdeep_kaur64@yahoo.co.in
DOI : http://dx.doi.org/10.13005/bbra/2643
ABSTRACT: Two isolates of Azotobacter chroococcum (A. chroococcum CBD15 and A. chroococcum W5), were characterized for their atmospheric nitrogen fixing efficiency and ability to produce plant growth promoting hormone. The isolates, A. chroococcum CBD15 and A. chroococcum W5, were observed the production of Indole acetic acid (IAA) and nitrogen fixation in the absence of any inorganic nitrogen source. The ability nitrogen fixation was estimated by acetylene reduction studies revealed that A. chroococcum CBD15 produced 693.3 nmole C2H4 h-1 mg-1 whereas A. chroococcum W5 produced 523.4 nmole C2H4 h-1 mg-1. Nitrogenase activity of both the isolates was reduced when grown in media containing nitrogen source (ammonia or urea), in comparison to media lacking any nitrogen source. The nifL gene, which is one of the most important regulatory gene of nitrogen fixation pathway, was isolated from A. chroococcum CBD15 and A. chroococcum W5. Sequence analysis revealed that both nifL gene sequences have maximum homology with nifL gene of A. vinelandii and Pseudomonas oryzae respectively. The genetic manipulation of nifL gene of A. chrococcum will lead to development of an efficient bioinoculant for sustainable agriculture.
KEYWORDS: Acetylene Reduction; Azotobacter Chroococcum; IAA; NifL; NifA
Download this article as:| Copy the following to cite this article: Sellamuthu G, Shankar P, Soundarapandian S, Paul S, Padaria J. C. Molecular Evolution of the Negative Regulatory Gene (NifL) from Azotobacter Chroococcum and its Nitrogenase Activity. Biosci Biotech Res Asia 2018;15(2). |
| Copy the following to cite this URL: Sellamuthu G, Shankar P, Soundarapandian S, Paul S, Padaria J. C. Molecular Evolution of the Negative Regulatory Gene (NifL) from Azotobacter Chroococcum and its Nitrogenase Activity. Biosci Biotech Res Asia 2018;15(2). Available from: https://www.biotech-asia.org/?p=30263 |
Introduction
Nitrogen is the most abundant and inert gas, present in the atmosphere and ‘nitrogen’ element is one of the most essential requirements for healthy plant growth. In natural systems, nitrogen required for plant growth comes from various sources as soil, rainfall, atmospheric deposition or biological nitrogen fixation. Plants can absorb nitrogen only in the form of ammonia or nitrate but the ammonia and nitrate levels are often very low in the soil. Thus, plants face poor nutritional availability with respect to nitrogen, causing negative effect on yield and higher nitrogen application timing did not affect the grain yield [1]. A few prokaryotic organisms have the nitrogen fixing ability ie they convert atmospheric nitrogen to a form that plants can easily uptake it. Biological nitrogen fixers are known to exert indirect effects on soil microbiological activities which in turn help the plant to grow better[2].
Among the prokaryotic nitrogen fixers, the most prominent are the heterotrophic diazotroph present in the rhizosphere of plants. These prokaryotes are capable of utilizing atmospheric nitrogen for their own growth; and as products of nitrogen fixation process, they also release soluble forms of nitrogen in the soil [3, 4]. They exist as free-living or as symbionts associated with cyanobacteria, various plants including grasses, woody plants and a number of legumes in diverse habitats of soil and water [5].
The nitrogen fixing microbes such as Klebsiella pneumoniae and Azotobacter vinelandii was NEshed?)well studied. Members of the genus Azotobacter belong to the family Azotobacteriaceae and are free-living obligately aerobic, nitrogen fixing heterotrophic diazotrophs. They include gram-negative γ-proteobacteria and exist as rod-shaped cells varying in size from 2.0-7.0 to 1.0-2.5 μm (length X breadth). A. chroococcum was the first species of Azotobacter genus to be described followed by other species which include A. vinelandii, A. armeniacus, A. paspali, A. beijerinckii, A. nigricans and A. salinestris [6].
The complex biological process of nitrogen fixation is governed by a number of enzymatic complexes. One of the key enzymes that crucial role in catalyzing biological nitrogen fixation is nitrogenase. Nitrogenase is a metalloenzyme that subdivides two components, which called their related to presence of metal irons such as Fe-protein and MoFe-protein [5]. Iron (Fe) protein plays important role in ATP-dependent electron donor to the larger component. It contains covalently linked two α subunits of metal cluster ([4Fe–4S] cluster). The [4Fe–4S] cluster involved in electron transfer to FeMo-protein which cycles between reduced and oxidized state [7]. The MoFe protein, contains the enzyme catalytic site [8]. The group of genes responsible for biological nitrogen fixation is together termed as ‘nitrogen fixation genes’ or nif genes. So far, twenty one nif genes have been reported from different nitrogen fixing bacteria. All the reports demonstrated that minimum of six conserved genes: nifH, nifD, nifK, nifE, nifN, and nifB genes were found in all diazotrophs for nitrogen fixation. The genes responsible for regulation of nitrogen fixation, nifH, nifD and nifK are considered as the most important nif genes of the nif operon as they together code for the metalloenzyme called nitrogenase. Nitrogenase plays a pivotal role in catalyzing biological nitrogen fixation along with the gene nifA, the positive regulator of the nif operon.
The NifA is an activator and positive regulator-encoding gene that binds to specific upstream gene sequence of nif genes to initiate transcription by interacting with σ54 subunit of the RNA polymerase. The expression of the gene nifA leads to expression of genes related to nitrogenase biosynthesis including the structural gene cassette nifHDK. Molecular biology techniques are been utilized for identification of nitrogen fixing bacteria based on the presence of structural gene (nifHDK) and other genetic elements. Sequence analysis of nifH gene has shown that like all other nif genes, nifH is also highly conserved across all nitrogen-fixing organisms indicating that this gene has not undergone many changes through evolutionary process [9]. Gene nifL acts as a negative regulator of the nif operon, in the presence of inorganic nitrogenous source as ammonia (fixed Nitrogen) [10]. Although there has been immense study on different biological nitrogen fixers yet we still need to identify an efficient microbial nitrogen fixer based on geographical location, agroclimatic zone and associative crop. Moreover, intensive molecular biology approaches are needed to understand and rectify the negative regulation of gene nifL [11].
The present study aimed at assessing the nitrogen fixing and growth promoting activity of two native isolates of A. chroococcum (A. chroococcum CBD15 and A. chroococcum W5) for their utilization as bioinoculants for enhanced nitrogen availability in soil for crop plants. Further, with the aim of manipulating the nifL gene for deleting its negative regulation ability, the nifL gene from both the isolates, A. chroococcum CBD15 and A. chroococcum W5 were sequenced and characterized.
Materials and Methods
Azotobacter Strains and Growth Medium
Azotobacter isolates (A. chroococcum CBD15 and A. chroococcum W5) were collected from Division of Microbiology, Indian Agricultural Research Institute, New Delhi [12]. The strains were further sub-cultured on Burk’s medium [13]. The purity of the cultures was checked by streak plate method and maintained on the slants of respective medium at 4°C in a refrigerator for medium term storage.
Molecular Characterization of nifL
Extraction of genomic DNA from the A. chrococcum isolates CBD15 and W5 was carried out by manual method [14]. The overnight cultures (OD= 1.2) were harvested and the pellet was washed with 0.7% NaCl. The pellet consisting of bacterial cells was treated with lysis solution, containing Tris (100mM), EDTA (10mM) and SDS (0.1%) and Proteinase K (20mg/l). The lysate was incubated at 65°C for 30 minutes to release nucleic acid. Following its release proteins and cellular debris removed by precipitation and chloroform: isoamyl alcohol (24:1) separation. The nucleic acid was then recovered from the clarified lysate by adding 0.7 volume of isopropanol. The purity of the nucleic acid was determined by measuring its optical density at 260nm and 280nm using Nano drop (Nanodrop 6000, Thermo Fisher). The nifL gene was amplified in A. chrococcum isolates CBD15 and W5 through polymerase chain reaction (PCR) using gene specific primers (forward F 5’-ATCGCGCTTTCCGCACCATCACCG-3’; reverse R 5’- TCGACTCTAGAGGTGAGGCCGA-3’) and genomic DNA of these isolates as template. The primers were designed based on sequence available in the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/). The PCR reaction mix contained in a final volume of 50μl: 5μl buffer (Genei), 1.5μl MgCl2 (50mM), 5μl dNTP-mix (2.5mM each), 1μl each primer (10μM), 10 ng purified DNA of cultured strains and 1U Taq-DNA polymerase (Genei). The PCR was performed with initial denaturation for 5 minutes at 94°C, 35 cycles with 30s 94°C, 30s 55°C, 1 minute at 72°C, and final elongation for 5 minutes at 72°C. The obtained amplicons of PCR were separated by 1.5% agarose gel electrophoresis. The amplified PCR product were cloned in pGEM-T Easy vector (Promega, USA) as per manufacturer’s protocol and sequenced and the obtained nucleotide sequences were determined with NCBI BLAST database (https://blast.ncbi.nlm.nih.gov/).
Sequence Alignment and Phylogenetic Analysis
The amplicon of ~ 1.4kb was obtained by PCR reaction. The resulting 1.4kb nucleotides sequence were BLAST searched. The new sequences were analyzed with those in the databases by using the basic local alignment search tool (BLAST; http://ncbi.nih.gov/cgi-bin/nph-blast?Jform=1). The sequences were aligned with selected genera with the CLUSTAL W program. The phylogenetic tree was analyzed with known sequence from NCBI gene bank from the neighbor-joining method with MEGA version 6.0 [15]. nifL sequence of A. chrococcum isolate CBD15 and A. chrococcum isolate W5 were submitted to GenBank to get the accession numbers.
Nitrogenase Activity Assay
Nitrogenase activity of the Azotobacter isolates was measured by acetylene reduction assay [16]. The isolates were inoculated in 5 ml of media and closed with rubber stoppers. The 5ml of air was replaced with 5 ml of acetylene gas (C2H2) using a sterile syringe and incubated for 24 hours at temperatures ranging between 28°C and 30°C. Following this standing period, 0.2 ml of gas injected into gas chromatography column to verify the amount of C2H2 converted to ethylene (C2H4) in each vial. To determine the amount of nitrogenase activity were following the formula used:
N = (hx × C ×V) / (hs × 24. 9 × t x mg protein)
Where, N = the concentration of C2H4 (nmol ml-1 h-1); hx = sample peak value: C = standard concentration of C2H4 (nmol ml-1h-1); V = volume of the vial; hs = standard peak value; t = reaction time (h).
The concentration of bacterial protein was determined by Lowry method [17]. To 0.2 ml of digested bacterial cell suspension was added to a total volume of 1 ml followed by addition of 5 ml of reagent “C” (copper sulphate-sodium-potassium tartrate solution) and immediate vortexing. Subsequently, the bacterial cell suspension was kept for 10 minutes at room temperature, and added 0.5 ml of reagent “D” (alkaline copper sulphate) and subsequently kept for incubation in the dark for 30 minutes. After 30 minutes of incubation, optical density of the reaction mixture was measured at 660nm and the amount of protein was determined with known concentration of standard protein (Bovine serum albumin).
Ammonia and Urea Sensitivity Assay
To assess the sensitivity of the Azotobacter strains to ammonia, the overnight cultures were inoculated in different media: Burk’s medium [13]. Burk’s medium+ ammonia (0.11%); Burk’s medium + 0.1% with urea. The cultures were incubated at 30˚C for 48 hours with 200rpm and the OD at 600 nm of culture was measured.
Indole Acetic Acid (IAA) Production Assay
The isolates, A. chrococcum CBD15 and A. chrococcum W5 were estimated the production of Indole Acetic Acid (IAA) using the protocol given by Bric et al. [18]. A. chrococcum isolates were inoculated in 100ml of ‘Nitrogen free’ media ie medium devoid of any nitrogen source. Another set of inoculations of A. chrococcum isolates was carried out in 100 ml of ‘Nitrogen free’ supplemented with tryptophan (100 µg ml-1). All the cultures were incubated at 30°C with shaking of 120 rpm (Kuhner LT-X (Lab-Therm) Switzerland) in for seven days. Subsequently, 2 ml of the liquid culture was transferred to centrifuge tube and centrifuged at 10,000 rmp (Eppendorf™ Model 5810R, Germany) for 15 minutes. The supernatant (1ml) transferred to a fresh centrifuge tube and mixed with 100μl orthophosphoric acid and Salkowski’s reagent (1 ml of 0.5 M FeCl3 in 50 ml of 35% HClO4) each and incubated up to colour formation. The colour absorbance was measured spectrophotometrically at 530 nm.
Statistical Analysis
All the experimental results were subjected to analysis of variance (ANOVA) and significance at 5% level was tested by Least Significant Difference (LSD) using SAS package, Version 8.2 (SAS version 8.2, 2001). (https://support.sas.com/downloads/browse.htm?cat=84).
Results
Molecular Characterization of nifL
The DNA was isolated from A.chroococcum CBD15 and A.chroococcum W5 cultures and PCR carried out for gene specific amplification of nifL gene using primers designed based DNA sequence of A. vinelandii gene nifL obtained from NCBI database (https://blast.ncbi.nlm.nih.gov/) The nifL amplicons of ~1.4kb obtained (Fig.1) in both the isolates were cloned in pGEMT vector and sequenced. The sequence analysis revealed that amplicons of 1416bp and 1303bp were obtained in A. chroococcum CBD15 and A. chroococcum W5 respectively which was confirmed as partial sequence of nifL gene.
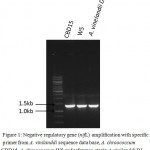 |
Figure 1: Negative regulatory gene (nifL) amplification with specific primer from A. vinilanddi sequence data base, A. chroococcum CBD15, A. chroococcum W5 and reference strain A. vinilanddi DJ. |
Sequence Alignment and Phylogenetic Analysis
Nucleotide sequence analysis of isolates A. chroococcum CBD15 and A. chroococcum W5 using ClustalW program revealed that both the isolates showed maximum homology (98%) with A. vinelandii and Pseudomonas oryzae (Fig. 2). An unknown sequence profile was constructed from the conserved region of negative regulatory gene from A. chrococcum isolates CBD15 and W5 with the identical nifL gene sequence from NCBI data base using ClustalW program. The results show that nifL gene of A. chroococcum CBD15 and A. chroococcum W5 were highly similar in sequence (99%, W5:0.00000, CBD15:0.00073). Similarity analysis of gene nifL of A. chroococcum CBD15 and A. chroococcum W5 with gene nifL of A. vinelandii (X64832.1) and A. chroococcum (CP010415.1) shows that percent identity matrix was 0.04054 (CP010415.1) and 0.00160 (X64832.1) Yellow and Green color highlighted (Fig 2a).
 |
Figure 2: Phylogenetic trees for Negative regulatory gene (nifL) showing the phylogenetic position of A. chroococcum among all known corresponding sequences of various diazotrophs. Trees were constructed by the neighbour-joining method. |
 |
Figure 2a: Homologies between the nifL gene ot A. chroococcum of CBD15 and W5 and other negative regulatory gene from A. vinilanddi and A. chroococcum. |
Homology to A. chroococcum of CBD15 and W5 nifL is shown highly identical with A. vinilanddi (pink), A. chroococcum of CBD15 and W5 differ from other A. vinilandii and A. chroococcum (green).
GenBank/EMBL Accession Number
Accession number for nifL sequence of A. chrococcum isolates CBD15 and A. chrococcum isolates W5 was KY781893 and MF375754 respectively.
Nitrogenase Activity Assay
The nitrogenase activity of A. chroococcum CBD15 was found to be the highest under aerobic conditions as compared to the A. chroococcum W5 with respect of all three different medium compositions used in the study (Fig. 3). The highest nitrogenase activity of A. chroococcum CBD15 was found in media devoid of any nitrogen source (693.3 nmole C2H4 h-1 mg-1), However, in media containing 0.1% urea, there was a decrease in nitrogen fixed (365.0 nmole C2H4 h-1 mg-1). With 0.11% of ammonium acetate as nitrogen source in the media, nitrogen fixation ability of A. chroococcum CBD15 further reduced (213.2 nmole C2H4 h-1 mg-1) (Fig. 3). Similarly in the case of A. chroococcum W5 nitrogenase activity was found to be higher in media without nitrogen source (523.4 nmole C2H4 h-1 mg-1) and decreased with increase in nitrogenous source ( 0.1% urea :354.9 nmole C2H4 h-1 mg-1; 0.11% ammonium acetate: 204.4 nmole C2H4 h-1 mg-1) (Fig. 3).
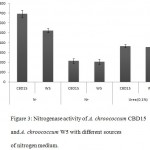 |
Figure 3: Nitrogenase activity of A. chroococcum CBD15 and A. chroococcum W5 with different sources of nitrogen medium. |
Error bars refer to standard deviation by means of five replicates.
Ammonia and Urea Sensitivity Assay
The growth characteristics of isolates A. chroococcum CBD15 and A. chroococcum W5 were determined. The isolates were grown in two different media having different supplementation of nitrogen sources (0.1% urea and 0.11% ammonium acetate). The growth of A. chroococcum CBD15 was observed to gradually increase in media containing 0.11% ammonium as was evident by OD at 600nm of the culture which increased from 0.23 to 0.66 in 48 hours. However, no changes were observed in the growth of A. chroococcum CBD15 in medium devoid of any nitrogen source, even after 48 hours. It was observed that in media with 0.1% urea, the growth of culture was very rapid during initial period as evident by the increase in OD at 600nm from 1.45 to 1.56 within 24 hours, however, as the time interval lengthened; the OD at 600nm of the culture began to decrease from 1.56 to 1.12 in 48 hours. Similarly observations were also made in case of growth of A. chrococcum W5 in media supplemented with different nitrogen sources. The growth of A. chroococcum W5 gradually increased in media containing 0.11% ammonium as was evident by OD at 600nm of the culture which increased from 0.23 to 0.66 in 48 hours as it was in case of A. chroococcum CBD15. With 0.1% urea in the media, the growth of culture was very rapid during initial period as evident by the increase in OD at 600nm from 1.34 to 1.78 within 24 hours, however, as the time interval lengthened; the OD at 600nm of the culture began to decrease from 1.42 to 1.18 in 48 hours (Fig. 4).
 |
Figure 4: Growth curve of A. chroococcum CBD15 and A. chroococcum W5 with different sources of nitrogen supplemented with medium.
|
Error bars refer to standard deviation by means of five replicates.
Indole Acetic Acid (IAA) Production Assay
The IAA production was measured depending on growth phase of bacterial isolates and maximal production of IAA was found in stationary phase. Since presence of tryptophan is necessary for IAA production, the optimum IAA production was achieved when medium was supplemented with tryptophan (100 µg ml-1). IAA production in presence of tryptophan (100µg ml-1) in both the isolates with A. chroococcum CBD15 and A. chroococcum W5 producing 16.9 µg ml-1 and 16.6 µg ml-1 of IAA respectively (Fig. 5). There is no distinct differences were observed in IAA production in presence of tryptophan. However, in medium lacking tryptophan, IAA production drastically reduced in A. chroococcum CBD15 (4.4 µg ml-1) and A. chroococcum W5 (4.2 µg ml-1).
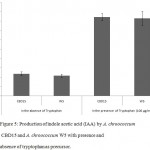 |
Figure 5: Production of indole acetic acid (IAA) by A. chroococcum CBD15 and A. chroococcum W5 with presence and absence of tryptophan as precursor. |
Error bars refer to standard deviation by means of five replicates.
Discussion
Several factors as excess ammonia, oxygen and temperature as well as other factors of the soil properties are reported to adversely affect the process of nitrogen fixation carried out by the soil microbial communities. The expression of nif genes for biological nitrogen fixation required functional nitrogenase. In our study, we aimed to characterize two isolates of A. chroococcum (A. chroococcum CBD15 and A. chroococcum W5) with respect to nitrogen fixing ability and plant growth promoting (PGP) hormone production. Therefore, the sequence analysis of the gene nifL was carried out in both the A. chrococcum isolates under study so as to understand the molecular organization of the nifL gene in A. chroococcum and to establish its relationship with the already well studied nifL gene from A. vinelandii.
Phylogenetic analyses of nifL sequence revealed that isolates of A. chroococcum, CBD15 and W5 shared maximum homology (98%) with A. vinelandii with and Pseudomonas oryzae and are thus closely related to them. BLAST database to determine the unknown sequence data by comparing query sequences which already reported greater number of sequences in public domain at EMBL, NCBI and DDBJ, as well as GenBank database that contains more than 260000 nucleotide sequences for organisms [19, 20]. The most conserved region of negative regulatory element in dizotophic bacteria and worldwide accepted for phylogenetic analysis and classification of bacterium, to serving as molecular chronometer. Similarly, reported that partial nifH gene sequences provided the information on the classification of phylogeny of diazotroph natural communities [21] phylogenetic tree of nifH gene from Paenibacillus sabinae [22]. Therefore, molecular characterization of genes is important get reliable and accurate identification of bacterial strains. The role of gene nifL in negative regulation of nif operon and ultimately nitrogen fixation pathway is well defined [23, 24]. The identification of closely related species was performed BLAST search. The bootstrap values ranged from 60- 100% which indicates Azotobacteracae family (denoted at the node). Azotobacter was the first genus reported that having alternative nitrogenases: vnfH and anfH [25]. The similar reports of nifH gene of Paenibacillus azotofixans NifH1 and NifH2 clustered with Cyanobacterium, P. azotofixans NifH3 clustered with NifH proteins of Archaea. P. sabinae NifH1, NifH2 and NifH3 clustered with Cyanobacterium. The nifH1, nifH2 and nifH3 of P. sabinae have 52–95% identities with nifH1, nifH2 and nifH3 of P. azotofixans, respectively. The identities of nifH1, nifH2 and nifH3 of P. sabinae with nifH1, nifH2, nifH3, nifH4, nifH5 and nifH6 of C. pasteurianum range between 56% and 60%, respectively [22]. The NifL protein regulate the expression of nitrogen fixation (nif) genes in presence of both the oxygen and fixed nitrogen or available nitrogen sources status. The negative regulator protein (NifL) decreased the expression of NifA, which transcriptional activator belonging to the bacterial enhancer-binding protein family. NifA is the gene product of nifA which is known to be positive regulator of nif operon [26].
Nitrogen is a most essential element for crop improvement and yield through plant growth and development and forms an integral part of life cycle of all living forms on earth. The diazotrophic microorganisms through enzymatic nitrogen fixation play an important function in conversion of atmospheric nitrogen to soluble nitrogen in a form that becomes naturally available to plants and microbes in soil. The ability of converting acetylene to ethylene is an indirect measurement of nitrogen fixation which is the specific activity of nitrogenase enzyme, and is indicative of nitrogen fixing soil microbes [27, 28]. The nitrogenase activity of isolates of A. chroococcum in our study was observed to decrease when nitrogen source was available in the media. Nitrogenase activity in A. chroococcum CBD15 when grown in a nitrogen source devoid medium decreased from 693.3 nmole C2H4 h-1 mg-1 to 365.0 nmole C2H4 h-1 mg-1 and 213.2 nmole C2H4 h-1 mg-1 when grown in medium with 0.1% urea and medium with 0.11% of ammonium acetate respectively. Similarly in case of A. chroococcum W5 nitrogenase activity was found to be higher in media without nitrogen source (523.4 nmole C2H4 h-1 mg-1) in comparison to media with 0.1% urea (354.9 nmole C2H4 h-1 mg-1) and with 0.11% ammonia (204.4 nmole C2H4 h-1 mg-1) (Fig.3). Previous reports also confirm that nitrogenase activity of nitrogen fixing bacteria rapidly decreased as ammonium concentration increased from 1mM (with glutamine concentration constant at 1.3 mM) in media to 10mM, and was finally switched off at ammonium concentration of 0.1 mM. At 0.1 mM ammonium, the rapid and total loss of activity was reversible and a ‘switch-on’ was observed in Pseudomonas stutzeri [29]. A. vinelandii is also reported that expression of all nif genes was reduced in the presence of external source of nitrogen. The negative regulatory gene, nifL is present immediately upstream of gene nifA which is considered to be positive regulator of the cascade of nif genes [30]. Gene nifA was active in the absence of fixed nitrogen in the culture medium. Oliveira et al., [10] report that inactivation of NifA in response to fixed nitrogen, which transcriptionally regulated by σ54 transcriptional activator of nifA in Herbaspirillum seropedicae. The inhibitor NifL regulates NifA activity depending on the reduction status of its N-terminally bound FAD-cofactor and allows NifA expression under anaerobic conditions [31, 32]. However, nif gene expression is tightly controlled at the transcriptional level with respect to the fixed nitrogen concentration, external nitrogen source. The earlier studies in which nif cluster of Paenibacllus is subject to similar regulation in presence of ammonia and oxygen, Paenibacillus sp. WLY78 and the engineered E. coli 78-7 strain did not exhibit nitrogenase activity at oxygen and ammonia concentrations above 5 percent and above 1 mM respectively [33].
Under natural conditions, bacterial isolates have the ability to produce phytohormones and successfully inhibiting pathogens by production of antibiotics, siderophores. Indole-3-acetic acid (IAA) involves cell enlargement and division, tissue differentiation, and responses to light [34]. The present results are similar with other studies where Pseudomonas putida has been shown to produce IAA when grown in a media supplemented with tryptophan as precursor of IAA. However, in medium lacking tryptophan, IAA production drastically reduced in A. chroococcum CBD15 (4.4 µg ml-1) and A. chroococcum W5 (4.2 µg ml-1). Similarly our results were agreed with a number of bacteria with plant growth promotion property such as as Azospirillum sp., Enterobacter cloacae, Alcaligenes faecalis, Acetobacter diazotrophicus, Klebsiella sp., Rhizobium, Klebsiellaoxytoca and Pseudomonas putida [34]. The minimum inhibitory effect of inorganic nitrogen on growth of A. chroococcum CBD15 was evident as the growth performance of the isolates A. chroococcum CBD15 and A. chroococcum W5 improved as evident from OD600 which increased from 0.23 to 0.66 in 48 hours in presence of nitrogen sources (0.11% ammonium) in the medium.. However, no growth changes were observed in both the isolates when cultured in medium devoid of any nitrogen source, even after 48 hours. It was observed that there was rapid increase in growth of A. chrococcum CBD15 and A. chrococcum W5 within 24 hours when grown in media supplemented within 0.1% urea in the media as evident from OD at 600nm (Fig. 4). Mannitol used as carbon sources and isolated Azotobacter from plants rhizosphere [35]. Besides nitrogen source, micronutrients are also reported to influence the growth of A. chrococcum. Lu et al., [36] reported that A. chroococcum strains able to grow on a zinc-free medium as well as media containing a zinc concentration upto 6-40 ppm, they suggested that zinc concentration of 20ppm in soil was optimum for Azotobacter growth. Further studies in this direction with our isolates would prove useful in developing bioinoculants for biological nitrogen supply to crops.
In summary our results demonstrate that a negative regulatory gene (nifL) from an Azotobacter can inhibit the nitrogenase activity when trace amount of external nitrogen sources or available nitrogen. Also several reports were demonstrated deletion of nifL gene can overcome nif gene expression. It rises various questions which concerning the mutation for negative regulatory elements (nifL) for fixating nitrogen efficiently by engineering diazotophic eukaryotes through important biotechnological approach.
Acknowledgments
Authors are also thankful to the Project Director, ICAR-National Research Centre on Plant Biotechnology, New Delhi for providing facilities to carry out the research work. Authors are thankful to Department of Biotechnology (DBT), Govt. of India, for providing financial grant under “Design and construction of a strong promoter for constitutive overexpression of nifA gene in Azotobacter vinelandii” project. Financial assistance from ICAR-National Innovations in Climate Resilient Agriculture project, New Delhi in the form of Research Associate fellowship of the first author during the latter stage of study is duly acknowledged. Authors duly acknowledge Prof. H. K. Das for his guidance during these experiments. Authors duly thank Dr. Dolly Wattal Dhar, Department of Microbiology, IARI, New Delhi, for providing the GC instrument facility.
Conflict of Interest
The authors declare that they have no potential conflict of interest related to this study.
References
- Nascente A.S, Carvalho M, da Conceição S, Melo L.C, Rosa P.H. Nitrogen management effects on soil mineral nitrogen, plant nutrition and yield of super early cycle common bean genotypes. Acta Scientiarum. Agronomy., 2017; 39 (3) :369-378.
CrossRef - Singh A. Singh J.N. Studies on influence of bio fertilizers and bio regulators on flowering, yield and fruit quality of strawberry Cv. Sweet Charlie. Ann. Agric. Res, 2006;25: 261-264.
- Damir O, Mladen P.I, Božidar S, Sran N. Cultivation of the bacterium Azotobacter chroococcum for preparation of biofertilizers. African. J. Biotech., 2011; 10 (16):3104-3111.
CrossRef - Sayeda M.A, Mohamed I.A.W, Wafaa T.A. Evaluation of Azotobacter and Azospirillum biofertilizers as a probiotic in Oreochromis niloticus aquaculture. J. Fisheries Aquatic. Sci., 2011; 6:535-544.
- Dixon R, Kahn D. Genetic regulation of biological nitrogen fixation. Nature. Reviews. Microbiol., 2004; 2: 621-631.
CrossRef - Robert L.R, Robert J.R., Moyra R, Ariel. S, Toby H. Azotobacter Genomes: The Genome of Azotobacter chroococcum NCIMB 8003 (ATCC 4412). PLOS ONE., 2015; 2015: 1-35.
- Igarashi R.Y, Seefeldt L.C. Nitrogen fixation: the mechanism of the Mo-dependent nitrogenase. Crit. Rev. Biochem. Mol. Biol., 2003; 38:(4): 351-384.
CrossRef - Rees D.C, Akif Tezcan F, Haynes C.A, Walton M.Y, Andrade S, Einsle O, Howard J.B. Structural basis of biological nitrogen fixation. Philos. Transact. Math. Phys. Eng. Sci., 2005; 363:971–984; discussion 1035–1040.
- Zehr J.P, Jenkins B.D, Short S.M, Steward G.F. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol., 2003; 5:539–554.
CrossRef - Oliveira M.A, Aquino B, Bonatto A.C, Huergo L.F, Chubatsu L.S, Pedrosa F.O, Souza E.M, Dixon R, Monteiro R.A. Interaction of GlnK with the GAF domain of Herbaspirillum seropedicae NifA mediates NH4 + -regulation. Biochimie., 2012; 94:1041–1047.
CrossRef - Bageshwar U.K, Srivastava M, Pardha S.P, Paul S, Gothandapani S, Jaat R.S, Shankar P, Yadav R, Biswas D.R, Kumar P.A, Padaria J.C, Mandal P.K, Annapurna K, Das H.K. An environmentally friendly engineered Azotobacter strain that replaces a substantial amount of urea fertilizer while sustaining the same wheat yield. Appl. Environ. Microbiol., 2017; 83:e00590-17.
CrossRef - Paul S, Rathi M.S, Tyagi S.P. Interactive effect with AM fungi and Azotobacter inoculated seed on germination, plant growth and yield in cotton (Gossypium hirsutum). Indian J. Agri. Sci., 2011; 81(11):1041–1045.
- Kennedy C.R, Gamal R.H, Ramos K.B, Dean D. The nifH, nifM and nifN genes of Azotobacter vinelandii: characterization by Tn5 mutagenesis and isolation from pLARF1 gene banks. Mol. Gen. Genet., 1986; 205:318–325.
CrossRef - George G, Brenner D.J, Krieg N.R, James R. Staley, Bergey’s Manual of Systematic Bacteriology. vol. 2: The Proteobacteria, Part B: The Gammaproteobacteria. 2007; Springer Science & Business Media
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol.Biol. Evol., 2013; 30:2725-2729.
CrossRef - Dilworth M.J. Acetylene reduction by nitrogen-fixing preparations from Clostridium pasteurianunz. Biochimica. Et. Biophysica. Acta., 1966; 127:285-294.
CrossRef - Lowry O.H, Rosebrough N.J, Farr A.L, Randall R.J. Protein measurement with the Folin Phenol reagent. J. Biol. Chem., 1951; 93:265-275.
- Bric J.M, Bostock R.M, Silverstone S.E. Rapid in situ assay for indoleacetic-acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microb., 1991; 57::535–538.
- Andrew M. The use of 16S RDNA Methods in Soil Microbial Ecology. Braz. J. Microbiol., 2000; 3:77-82.
- Benson D.A, Karsch-Mizrachi Ilene, Lipman D.J, Ostell J. Wheeler D.L. GenBank. Nucleic Acids Research., 2008; 36:D25–D30.
CrossRef - Franck P, Ranjard L, Nazaret S, Gourbiere F, Monrozier L.J. Comparison of nifH Gene Pools in Soils and Soil Microenvironments with Contrasting Properties. Appl. Environ. Microbiol., 2001; 67:2255-2262.
CrossRef - Hong Y, Maa Y, Wua L, Maki M, Qinb W, Chen S. Characterization and analysis of nifH genes from Paenibacillus sabinae T27. Microbiological Research., 2012; 167:596–601.
CrossRef - Dixon R.A. The Genetic Complexity of Nitrogen Fixation, The Ninth Fleming Lecture. J. Gen. Microbio., 1984; 130:2745-2755.
- Poza-Carrión C, Jiménez-Vicente E, Navarro-Rodríguez M, Echavarri-Erasun C, Rubi L.M.. Kinetics of nif Gene Expression in a Nitrogen-Fixing Bacterium. J. Bacteri., 2014; 196(3):595-603.
CrossRef - Jianyin L, Mengjun P, Youguo L. Phylogenetic diversity of nitrogen-fixing bacteria and the nifH gene from mangrove rhizosphere soil. Can. J. Microbiol., 2012; 58:531–539.
CrossRef - Martinez-Argudo I, Little R, Dixon R. Role of the amino-terminal GAF domain of the NifA activator in controlling the response to the antiactivator protein NifL. Molecul. Microbiol., 2004;\52(6):1731–1744.
- Andrade G, Mihara K, Linderman R, Bethlenfalvay G.J. Bacteria from rhizosphere and hyphosphere soils of different arbuscular-mycorrhizal fungi. Plant Soil., 1997; 192:71.
CrossRef - Manoj K, Rajesh K. Acetylene reductase activity and molecular characterization of plant growth promoting rhizobacteria to know efficacy in integrated nutrient management system. Indian J. Biotech., 2015; 14:221-227.
- Desnoues N, Lin M, Guo X, Ma L, Carreño-Lopez R, Elmerich C. Nitrogen fixation genetics and regulation in a Pseudomonas stutzeri strain associated with rice. Microbiology., 2003; 149: 2251–2262.
CrossRef - Little R, Slavny P, Dixon R. Influence of PAS Domain Flanking Regions on Oligomerisation and Redox Signalling By NifL. PLoS ONE., 2012; 7(10): e46651.
CrossRef - Klopprogge K, Grabbe R, Hoppert M, Schmitz R.A. Membrane association of Klebsiella pneumoniae NifL is affected by molecular oxygen and combined nitrogen. Arch. Microbiol., 2002; 177:223–234.
CrossRef - Grabbe R, Schmitz R.A. Oxygen control of nif gene expression in Klebsiella pneumoniae depends on NifL reduction at the cytoplasmic membrane by electrons derived from the reduced quinone pool. Eur. J. Biochem., 2003; 270:1555-1566.
CrossRef - Wang L, Zhang L, Liu Z, Zhao D, Liu, X, Zhang B, Xie J, Hong Y, Li P, Chen S, Dixon R, Li J. Correction: A Minimal Nitrogen Fixation Gene Cluster from Paenibacillus sp. WLY78 Enables Expression of Active Nitrogenase in Escherichia coli. PLOS Genetics., 2013; 9(10).
CrossRef - Leveau J.H.J, Lindow S.E. Utilization of the plant hormone indole-3-acetic acid for growth by Pseudomonas putida strain 1290. Appl. Environ. Microbiol., 2005; 71:2365–2371.
CrossRef - Shrivastava U.P. Isolation and initial characterization of diazotrophic plant growth promoting rhizobacteria (PGPR) from rice rhizosphere of Parsa and Bara district of Nepal. Inter. J. Pharmacy Life Sci., 2013.; 4:(3):1-8.
- Lu C.Q, Huang B.L. Isolation and characterization of Azotobacteria from pine rhizosphere. Afr. J. Microbiol. Res., 2010; 4(12):1299–1306.

This work is licensed under a Creative Commons Attribution 4.0 International License.





