How to Cite | Publication History | PlumX Article Matrix
Raman Jasrotia1 , Jyotdeep K. Raina2
, Jyotdeep K. Raina2 , Minakshee Sharma2, Rakesh K. Panjaliya1
, Minakshee Sharma2, Rakesh K. Panjaliya1 , B. R. Kundal3
, B. R. Kundal3  and Parvinder Kumar1,2
and Parvinder Kumar1,2
1Department of Zoology, University of Jammu, Jammu, 180006, India.
2Institute of Human Genetics, University of Jammu, Jammu, 180006, India.
3Department of Neurology, Govt. Super Speciality Hospital, Jammu, India.
Corresponding Author E-mail: parvinderkb2003@yahoo.co.in
DOI : http://dx.doi.org/10.13005/bbra/2694
ABSTRACT: Disturbance in vascular functioning pathways has been related to pathophysiology of migraine. The present study investigated the role of MTHFR C677T and ACE I/D gene polymorphisms in migraine susceptibility within the population of Jammu province of J&K state. A sum of 252 subjects including 102 migraine patients and 150 non-migrainous unrelated healthy controls were enrolled for the present study. PCR-RFLP was performed for determining MTHFR gene variations. For detecting insertion/deletion in ACE gene PCR was performed. In case of MTHFR, ‘T’ allele (variant allele) and TT genotype (variant) was found to be present only in migraine patients but not in controls thereby suggesting its positive role in migraine pathophysiology. For ACE I/D polymorphism, higher frequency of DD genotype (32.35 % vs 15.3 %) and D allele (0.51 vs 0.4) were observed in patients than in controls. Logistic regression analysis revealed a significant association of ACE I/D polymorphism with risk of migraine. However, a direct link of MTHFR C677T polymorphism with migraine risk was not found.
KEYWORDS: Angiotensin Converting Enzyme; Genotype; Migraine, Methylenetetrahydrofolate Reductase; Polymorphism
Download this article as:| Copy the following to cite this article: Jasrotia R, Raina J. K, Sharma M, Panjaliya R. K, Kundal B. R, Kumar P. Relationship of MTHFR and ACE Gene Variations With Migraine Susceptibility: A Case-Control Study in the Population of North India (Jammu). Biosci Biotech Res Asia 2018;15(4). |
| Copy the following to cite this URL: Jasrotia R, Raina J. K, Sharma M, Panjaliya R. K, Kundal B. R, Kumar P. Relationship of MTHFR and ACE Gene Variations With Migraine Susceptibility: A Case-Control Study in the Population of North India (Jammu). Biosci Biotech Res Asia 2018;15(4). Available from: https://www.biotech-asia.org/?p=32643 |
Introduction
Migraine is a common neurovascular primary headache disorder marked by recurrent episodes and attacks of disabling head pain with additional symptoms such as queasiness, vomiting, photophobia, sonophobia and visual distortions in some sufferers (Silberstein, 2004; Stuart et al., 2012). Migraine has been identified by WHO amongst the top twenty principal causes of disability across the globe (WHO, 2001; Leonardi et al., 2005). It is known to affect about 12% of the general population with clear predominance of migraine attacks in women over men (Lipton and Bigal, 2005; Joshi et al., 2009). International Headache Society (IHS) has classified migraine into two major subdivisions viz., migraine without aura (MO) and migraine with aura (MA) (HCCIHS, 1988). In case of MA, the patients report a distinct phase of focal neural disturbances named as “aura” preceding or accompanying the headache while in MO such disturbances are not found (HCCIHS, 2004). The aetiology of migraine is complex and multifactorial, involving coordination of both genetic and environmental components (Piane et al., 2007; Kundal et al., 2016). The genetic component of migraine is polygenic in nature with a number of susceptibility genes that are known to influence the risk of migraine. The changes in the vascular endothelial functioning have been observed in migraineurs (Colson et al., 2007; Kundal et al., 2016). Therefore, genes which are known to be linked with vascular or endothelial functioning have become prime candidates that may be involved in migraine pathogenesis. Two important vascular genes that have been taken up in the present study are MTHFR and ACE.
MTHFR (Methylenetetrahydrofolate reductase) enzyme converts the substrate 5,10-MTHF (methylenetetrahydrofolate) into 5-MTHF (methylenetetrahydrofolate) which is the principal form of folate in blood circulation. In the process of re-methylation of homocysteine to methionine 5-MTHF acts as carbon donor. (Frosst et al., 1995; Heux et al., 2004; Raina et al., 2016 b). The human MTHFR gene has been mapped to chromosome 1 (1p36.3). A common variant of the MTHFR gene is C677T in which valine (val) replaces alanine (Ala) by mutation at codon position 222. It has been reported that the enzymatic activity of MTHFR gets reduced in persons with Val residue with the mean enzymatic activity of 30% in (TT) homozygous (Val/Val) state and 65 % in the (CT) heterozygous (Ala/Val) state (Frosst et al., 1995). This reduced MTHFR activity can lead to elevation in plasma homocysteine levels particularly when intake of folate is less (Frosst et al., 1995; Wald et al., 2002). The increased homocysteine level is linked with the elevation of risk of vascular diseases and neurological conditions such as migraine (Frosst et al., 1995; Raina et al., 2016 a, Raina et al., 2016 b).
Angiotensin converting enzyme (ACE) is a zinc metalloenzyme which functions in several tissues, including vascular and endothelial cells and is known for regulating blood pressure and electrolytic balance (Yang et al., 2006). In RAAS (rennin-angiotensin-aldosterone system) ACE is one of the chief enzyme which converts angiotensin I into angiotensin II, an effective vasopressor (Costerousse et al., 1997; Kundal et al., 2016). The ACE gene has been mapped to chromosome 17 (17q23.3). It has two polymorphic alleles, insertion (I), and the deletion (D) of 287 bp Alu sequence within the intron 16 (Sharma et al., 1998., Kundal et al., 2016). (DD genotype) is related with increased levels of circulating ACE than heterozygous (ID) and homozygous (II) genotype (Rigat et al., 1990). There has been several interesting results published reporting relationship of ACE I/D polymorphism with migraine. The Deletion allele of ACE gene reported as a strong risk factor for susceptibility of migraine (Paterna et al., 2000; Kowa et al., 2005; Lea et al., 2005).
Objectives
The impact of MTHFR and ACE gene variations has been studied across worldwide populations. However, results for the potential association of these two gene polymorphisms with the risk of migraine remain controversial as some studies depicted significant association while others showed no association at all. Taking into account the paucity of the data on the relationship of MTHFR and ACE gene polymorphisms with migraine in North Indian population of Jammu the present work was undertaken to investigate the likely association of MTHFR C677T and ACE I/D polymorphisms with migraine susceptibility. This is the first attempt to evaluate the involvement of gene polymorphisms of these two important candidate genes of vascular function with migraine risk of Jammu population.
Materials and Methods
Subjects
The subjects comprised of 102 migraine patients and 150 non-migrainous healthy unrelated controls. Among 102 migraine patients, 75 were sufferer of migraine without aura (MO) and 27 were patients of migraine with aura (MA). Identification of migraine patients were done according to the criterion and classification of the International Headache Society (HCCIHS 2004) by concerned medical practionner. The migraine patients were enrolled from Out Patient Department (OPD) of Neurology Department, Govt. Super Speciality Hospital, Jammu where as controls were recruited from premises of University of Jammu. A written consent was obtained from each patient and study was ethically approved by Animal and Human Experimentation Ethical Committee (AHEEC), University of Jammu.
Blood Sample Collection and DNA Isolation
3 ml of whole blood was taken from each study participants by venipuncture into EDTA coated vials. Phenol-Chloroform-Isoamyl alcohol method (Sambrook and Russel, 2001) with slight changes was used for the isolation of genomic DNA from stored blood samples. The qualitative analysis of isolated genomic DNA was done by 0.8% Agarose gel electrophoresis and the quantitative analysis was done by Spectrophotometry.
Determination of MTHFR C677T Polymorphism
Various genotypes of MTHFR gene for C677T polymorphism were obtained by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) as discussed previously by Raina et al. (2016b). The sequence for forward primer was 5’-TGA AGG AGA AGG TGT CTG CGG GA-3’, and the reverse primer was 5’-AGG ACG GTG CGG TGA GAG TG-3’. The PCR reaction component included 100 ng genomic DNA, 13 µl dH2O, 5X Flexi buffer, 200 µM dNTPs, 0.2 µM of each primer, 1.5 mM MgCl2 and 1.25 U Taq polymerase on Applied Biosystems (Veriti).
The obtained 198 bp length PCR products (Fig 1) was then given a restriction digestion with Hinf I restriction endonuclease. The products of restriction digestion were detected by electrophoresis usind 3.5 % agarose gel stained with ethidium bromide. The obtained digested products included 198 bp for wild (CC) genotype, 198, 175 and 23 bp for heterozygous (CT) genotype and 175 and 23 bp for mutant (TT) genotype. (Fig 2).
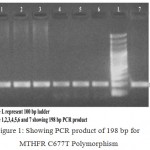 |
Figure 1: Showing PCR product of 198 bp for MTHFR C677T Polymorphism.
|
 |
Figure 2: Band sizes of MTHFR PCR product after restriction digestion with Hinf I.
|
Determination of ACE ID Polymorphism
Insertion-Deletion polymorphism of ACE gene was determined by polymerase chain reaction (PCR) using a pair of oligonucleotide primers as done by Kundal et al. (2016). The forward primer sequence was 5′-CTG GAG ACC ACT CCC ATC CTT TCT-3′ and reverse primer sequence was 5′-GAT GTG GCC ATC ACA TTC GTC AGA T-3′. The amplification products were obtained using reaction system containing100 ng genomic DNA, 13 µl dH2O, 5X Flexi buffer, 200 µM dNTPs, 0.2 µM of each primer, 1.5 mM MgCl2 and 1.25 U Taq polymerase on Applied Biosystems (Veriti).
The homozygous subjects (II) with insertion allele were recognized by the presence of a single 490 bp product fragment, while the homozygous (DD) for the deletion allele were recognized by single 190 bp product fragment and the heterozygous subjects (ID) with both insertion and deletion alleles were recognized by the occurrence of both 490 bp and 190 bp fragments. (Fig 3).
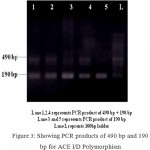 |
Figure 3: Showing PCR products of 490 bp and 190 bp for ACE I/D Polymorphism.
|
Statistical Analysis
In order to evaluate the significant difference between observed and expected genotypes among migraine cases and controls Hardy Weinberg equilibrium calculations were done. The association of MTHFR C677T and ACE ID polymorphisms with migraine susceptibility was analyzed by calculating the odds ratio (OR) with 95% confidence interval (CI). A p-value <0.05 is considered as statistical significant. Statistical analysis was completed by means of Statistical Package for Social Sciences (SPSS) software version 20.
Results
MTHFR C677T Polymorphism
Genotypic frequencies among patients were not in agreement with Hardy Weinberg equilibrium. Heterozygous (CT) and variant (TT) genotypes were absent among controls. However, the overall frequency of T allele in patients was found to be 0.02. The genotypic and allelic distribution for MTHFR C677T polymorphism was shown in Table 1. Due to absence of heterozygous and homozygous variant genotypes in control group, we were not able to apply genetic models for evaluating risk or protection conferred by genotype combinations towards migraine progression. However, we found that T allele was present only in patients indicating that the allele might have a positive role in migraine development especially in migraine without aura (Table 2). The different genotypes obtained on the whole in patients (cases) and controls depicted in Table 3.
Table 1: Genotypic and allelic frequency distribution in Migraine patients and Controls for MTHFR gene C677T Polymorphism.
| Category | Genotypic frequency (%) | Allelic frequency | Chi-square | p- value | |||
| CC | CT | TT | C | T | |||
| Cases (N=102) | 98
(96.08 %) |
3
(2.94 %) |
1
(0.98 %) |
0.98 | 0.02 | 15.11 | <0.001* |
| Controls (N=150) | 150
(100 %) |
0 | 0 | 1 | 0 | – | – |
N = Number, CC= Homozygous Wild, TT= Homozygous Mutant, CT = Heterozygous, * Significant value
Table 2: MTHFR C677T Polymorphism in MO patients.
| Genotypes/Alleles | Migraine without aura (MO) (N=75) | Controls (N=150) |
| CC | 71 | 150 |
| CT | 3 | 0 |
| TT | 1 | 0 |
| CT+TT | 4 | 0 |
| C | 0.97 | 1 |
| T | 0.03 | 0 |
Table 3: MTHFR C677T Polymorphism in Migraine patients in general.
| Genotypes/Alleles | Cases (N=102) | Controls (N=150) |
| CC | 98 | 150 |
| CT | 3 | 0 |
| TT | 1 | 0 |
| CT+TT | 4 | 0 |
| C | 0.98 | 1 |
| T | 0.02 | 0 |
ACE I/D Polymorphism
The genotypic and allelic allocation of study participants were presented in Table 4. The genotypic frequencies were in accordance with Hardy Weinberg equilibrium among control group whereas a significant deviation from Hardy Weinberg equilibrium was found in patient group. Overall, frequency of risk (D) allele was higher in patients (0.51) as compared to controls (0.4).
Table 4: Genotypic and allelic frequency distribution in Migraine patients and Controls for ACE gene I/D polymorphism.
| Group | Genotypic frequency (%) | Allelic frequency | Chi-square | p- value | |||
| II | ID | DD | I | D | |||
| Cases (N=102) | 30
(29.41% ) |
39
(38.24%) |
33
(32.35 %) |
0.49 | 0.51 | 5.62 | <0.05* |
| Controls (N=150) | 53
(35.3 %) |
74
(49.4 %) |
23
(15.3 %) |
0.6 | 0.4 | 0.12 | >0.05 |
N = Number, I = Insertion allele, D = Deletion allele, * Significant value
Logistic regression analysis for genotype combinations were given in Table 5, 6 and 7. Among sub-groups of migraine patients, significant differences were observed in the frequency of DD genotype in MO when compared with control group (OR=2.76; 95% CI=1.28–5.96; p =0.008).
Table 5: Association of ACE I/D Polymorphism with MO risk.
| Genotypes/Alleles | Migraine without aura (MO) (N=75) | Controls (N=150) | OR (95 % C.I) | p-value |
| II | 20 | 53 | 1(Reference) | |
| ID | 31 | 74 | 1.11 [0.57-2.15] | 0.7 |
| DD | 24 | 23 | 2.76 [1.28-5.96] | 0.008* |
| ID+DD | 55 | 97 | 1.50 [0.81-2.77] | 0.1 |
| I | 0.47 | 0.6 | 1(Reference) | |
| D | 0.53 | 0.4 | 1.7 [1.12-2.47] | 0.010* |
N = Number, OR = Odds ratio, CI = 95% Confidence interval, * Significant value
Table 6: Association of ACE I/D Polymorphism with MA risk.
| Genotypes/Alleles | Migraine with aura (MA (N=27) | Controls (N=150) | OR (95 % C.I) | p-value |
| II | 10 | 53 | 1(Reference) | |
| ID | 8 | 74 | 0.57 [0.21-1.54] | 0.27 |
| DD | 9 | 23 | 2.07 [0.74-5.78] | 0.16 |
| ID+DD | 17 | 97 | 0.92 [0.39-2.17] | 0.86 |
| I | 0.52 | 0.6 | 1(Reference) | |
| D | 0.48 | 0.4 | 1.39 [0.77-2.49] | 0.26 |
N = Number, OR = Odds ratio, CI = 95% Confidence interval,
Table 7: Association of ACE I/D polymorphism with Migraine risk in general.
| Genotypes/Alleles | Cases (N=102) | Controls (N=150) | OR (95 % C.I) | p-value |
| II | 30 | 53 | 1(Reference) | |
| ID | 39 | 74 | 0.93 [0.51-1.68] | 0.8 |
| DD | 33 | 23 | 2.5 [1.26-5.08] | 0.008* |
| ID+DD | 72 | 97 | 1.31 [0.76-2.25] | 0.3 |
| I | 0.49 | 0.6 | 1(Reference) | |
| D | 0.51 | 0.4 | 1.6 [1.11-2.27] | 0.01* |
N = Number, OR = Odds ratio, CI = 95% Confidence interval, * Significant value
The risk allele D was observed to give nearly 1.7 folds risk towards MO in the studied population. However, no significant differences were seen in frequency of DD genotype and D allele in MA when compared to control group as shown in Table 6.
Combined analysis of migraine patients with ACE I/D polymorphism was given in Table 7. OR analysis revealed that DD vs II genotype and D vs I allele combination was providing approximately 2.5 folds and 1.6 folds risk for migraine susceptibility in population of Jammu region. Furthermore, it could be speculated that ACE I/D polymorphism had a significant role in development of MO but not MA in studied population.
Discussion
Migraine is one of the commonest neurological disorders that puts a significant burden on society socially as well as financially. Its aetiology is complex and usually involves alteration in vascular functioning. Thus, genes involved in vascular functioning become the prime candidates that are invoved in migraine pathogenesis. The aim of this study was to find the relationship between MTHFR and ACE gene polymorphisms and migraine in the population of Jammu region.
MTHFR is the critical enzyme of homocysteine metabolism and is invoved in conversion of 5, 10-MTHF to 5-MTHF. (C to T) polymorphism in the MTHFR gene at nucleotide position 677 is responsible for the synthesis of thermolabile MTHFR enzyme with reduced enzymatic activity which causes to a moderate elevation of homocysteine level in blood plasma (Frosst et al., 1995; Wald et al., 2002). A strong and positive association between the MTHFR C677T variant and susceptibility of migraine has been reported by several earlier studies (Kowa et al., 2000; Kara et al., 2003; Lea et al., 2004; Oterino et al., 2004; Scher et al., 2006; Tietjen et al., 2009; Gavgani and Hoseinian, 2012; Bahadir et al., 2013). However, various other studies showed no association between MTHFR C677T polymorphism and migraine (Kaunisto et al., 2006; Todt et al., 2006; Ferro et al., 2008; Schurks et al., 2008; Rubino et al., 2009; Liu et al., 2010).
Since, in the present study we didnot get a significant association of MTHFR C677T polymorphism with migraine due to absence of CT and TT genotypes in control population. But the results of the present study showed higher frequency of CT and TT genotypes as well as T- allele in patient group thereby suggesting a possible association of T allele with disease outcome. In a study done in Chinese population, it was observed that the frequency of T allele was significantly higher in migraine without aura (MO) than in controls (An et al., 2013), which was compatible with present investigation. Also another study done on population of North India failed to establish an association of MTHFR C677T variant with migraine susceptibility (Joshi et al., 2009; Kaur et al., 2018). An investigation on Kashmiri population done by Pandith et al., (2017) for MTHFR gene C677T polymorphism also showed no role in predisposition to the migraine. Likewise our results, prior reports on MTHFR C677T polymorphism in Jammu region were also in support of lower prevalence of CT genotype and lack of TT genotype in control population (Raina et al., 2016 b). In another study on cardiovascular disorders in Jammu region (Raina et al., 2016 a) also reported complete absence of CT and TT genotypes in female control group. The findings of the present work were in concordance with previous studies done in population of Jammu region. Overall, we found higher prevalence of CC genotype and C allele in study population of Jammu region.
(ACE) is one of the chief components of RAAS which is involved in transformation of angiotensin I to the vasoactive angiotensin II (Rigat et al., 1990), thus modulating vascular tension and blood pressure. An elevation in circulating ACE levels may disturb neurovascular activity due to the alteration in the levels of neurotransmitters (Kowa et al., 2005). The DD genotype of ACE gene is found usually associated with is also associated with he raised frequency of attacks of migraine in patients of MO (Paterna et al., 2000). Similarly, Pizza et al., (2013) found higher frequency of DD genotype i.e. 50% in migraine patients which was significantly higher than that observed in controls 34% in Italian population. The presence of D allele has been linked to both MO and MA where as other researchers have shown its association with MA alone (Kowa et al., 2005; Lea et al., 2005). A case-control study from Taiwan did not reveal any differences in ACE allelic frequencies between migraine cases and controls, but strikingly the ACE-DD variant conferred slight protective effect against migraine in male patients (Lin et al., 2005). In our study, we establish that DD genotype and D allele of ACE gene was significantally associated with migraine in general and migraine without aura (MO) in particular as compared to healthy controls in the North population of Jammu province of J&K. These findings were in conformity with previous study done in Italian population by Paterna et al., (2000) in which the higher prevalence of DD genotype (48.34%) was found in MO patients than that of controls (37.32%) suggesting a significant association (p < 0.05) of ACE DD genotype with migraine without aura (MO). In an another study on North-Indian population have shown higher frequency of DD genotype (12%) in migraine patients when compared to healthy controls (8%), but the study failed to generate a significant relationship between DD genotype and migraine. However, it showed a significant association of DD genotype with MA [II vs DD, OR=0.04, 95% CI=1.045-7.662, p=0.04] (Joshi et al., 2009) which was contradictory to our results. In contrast to our results, some previous studies have shown no association of ACE ID polymorphism with migraine (Alicakmak et al., 2003; Tronvik et al., 2008; Alasehirli et al., 2009; Ozbey et al., 2010; Sezer et al., 2013).
Conclusion
The present communication has reported the presence of MTHFR 677 CT genotype and variant T allele only in cases but not in controls which suggests a possible involvement of variant T allele in migraine development whereas ACE I/D polymorphism analysis revealed that D allele and DD genotype is considerably associated with the risk for migraine in population of North India (Jammu region). The data generated in the present research work will serve as baseline for conducting further candidate gene studies on migraine susceptibility on larger sample size.
Acknowledgements
The authors would like to extend their gratitude to all the patients and volunteers involved in the study. The authors are also thankful to Prof. Seema Langer, Head, Department of Zoology, University of Jammu for providing necessary lab facilities.
Conflict of Interest
There is no conflict of interest.
Funding Sorce
The study was funded by J&K State Council for Science and Technology
References
- Alasehirli B., Gur M., Akçali A., Geyik S., Bulbul B., Sayar D., Yılmaz M., Neyal A and Neyal M. Angiotensin Converting Enzyme Gene Insertion/Deletion Polymorphism in Migraine Patients. Turk Norol Derg. 2009;15:161-165.
- Alicakmak., Cataloluk O., Yoldas T., Arslan A., Herken H., Barlas O., Gorucu S and Yigiter R. Migraine and angiotensin-converting enzyme association in Turkish patients. The Pain Clinic. 2003;15(4):473-477.
- An X. K., Lu C. X., Ma Q. L and Zhang X. R., Burgunder J. M., Lin Q., Qu H. L. Association of MTHFR C677T Polymorphism with Susceptibility to Migraine in the Chinese population. Neuroscience Letters. 2013;549:78–81.
- Bahadir A., Eroz R and Dikiei S. Investigation of MTHFR C677T Gene Polymorphism, Biochemical and Clinical Parameters in Turkish Migraine Patients: Association with Allodynia and Fatigue. Cell Mol Neurobiol. 2013;33(8):1055-63.
- Colson N. J., Fernandez F., Lea R. A and Griffiths L. R. The search for migraine genes: an overview of current knowledge. Mol. Life Sci. 2007;64:331–344.
- Costerousse O., Danilov S and Alhenc-Gelas F. Genetics of angiotensin I-converting enzyme. Exp. Hypertens. 1997;19(5-6):659-669.
- Ferro A., Castro M. J., Lemos C., Santos M., Sousa A., Pereira-Monteiro J., Sequeiros J and Maciel P. The C677T polymorphism in MTHFR is not associated with migraine in Portugal. Dis Markers. 2008;25:107–113.
- Frosst P., Blom H. J., Milos R., Goyette P., Sheppard C. A., Matthews R. G., Boers G. J., den Heijer M., Kluijtmans L. A and van den Heuvel L. P. A candidate genetic risk factor for vascular disease a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10(1):111-113.
- Gavgani S. C and Hoseinian M. M. Comparative study on homocysteine levels in migraine patients and normal peoples. Ann Bio Res. 2012;3:1804-1807.
- Headache Classification Committee of the International Headache Society. Classification and diagnostic criteria for headache disorders, cranial neuralgias and facial pain Ist edition. Cephalalgia. 1988;8(7):1–96.
- Headache Classification Committee for the International Headache Society. Classification and diagnostic criteria for headache disorders, cranial neuralgias and facial pain 2nd edition. Cephalgia. 2004;24( 1):1–60.
- Heux S., Morin F., Lea R. A., Ovcaric M., Tajouri L and Griffiths L. R. The methylentetrahydrofolate reductase gene variant (C677T) as a risk factor for essential hypertension in Caucasians. Hypertens Res. 2004;27:663-667.
- Joshi G., Pradhan S and Mittal B. Role of the ACE ID and MTHFR C677T polymorphisms in genetic susceptibility of migraine in a north Indian population. J Neurol Sci. 2009;277:133-137.
- Kara I., Sazci A., Ergul E., Kaya G and Kilic G. Association of the C677T and A1298C polymorphisms in the 5,10 methylenetetrahydrofolate reductase gene in patients with migraine risk. Brain Res Mol Brain Res. 2003;111:84–90.
- Kaur S., Ali A., Pandey A. K and Singh B. Association of MTHFR gene polymorphisms with migraine in North Indian population. Neurological Sciences. 2018;39:691–698.
- Kaunisto M. A., Kallela M., Hamalainen E., Kilpikari R., Havanka H., Harno H., Nissila M., Sako E., Ilmavirta M., Liukkonen J., Teirmaa H., Tornwall O., Jussila M., Terwilliger J., Farkkila M., Kaprio J., Palotie A and Wessman M. Testing of variants of the MTHFR and ESR1 genes in 1798 Finnish individuals fails to confirm the association with migraine with aura. Cephalalgia. 2006;26:1462–1472.
- Kowa H., Fusayasu., Ijiri T., Ishizaki K., Yasui K., Nakaso K., Kusumi M., Takeshima T and Nakashima K. Association of the insertion/deletion polymorphism of the angiotensin I-converting enzyme gene in patients of migraine with aura. Neurosci Lett. 2005;374(2):129–31.
- Kowa H., Yasui K., Takeshima T., Urakami K., Sakai F and Nakashima K. The Homozygous C677T Mutation in the Methylenetetrahydrofolate Reductase Gene Is a Genetic Risk Factor for Migraine. American Journal of Medical Genetics (Neuropsychiatric Genetics). 2000;96:762–764.
- Kundal B. R., Jasrotia R., Raina J. K., Bhardwaj R., Panjaliya R. K and Kumar P. Angiotensin Converting Enzyme (ACE) I/D Gene Polymorphism in susceptibility of Migraine. Indian Journal of Applied Research. 2016:6(6):57-59.
- Lea R. A., Ovcaric M., Sundholm J., MacMillan J and Griffiths L. R. The methylenetetrahydrofolate reductase gene variant C677T influences susceptibility to migraine with aura. BMC Med. 2004;2:3.
- Lea R. A., Ovcaric M., Sundholm J., Solyom L., MacMillan J and Griffiths L. R. Genetic variants of angiotensin converting enzyme and methylene tetrahydrofolate reductase may act in combination to increase migraine susceptibility. Brain Res Mol Brain Res. 2005;136(1–2):112–7.
- Leonardi M., Steiner T. J., Scher A. T and Lipton R. B. The global burden of migraine: measuring disability in headache disorders with WHO’s Classification of Functioning, Disability and Health (ICF). J Headache Pain. 2005;6(6):429-40.
- Lin J. J., Wang P. J., Chen C. H., Yueh K. C., Lin S. Z and Harn H. J. Homozygous deletion deletion genotype of angiotensin converting enzyme confers protection against migraine in man. Acta Neurol Taiwan. 2005;14(3):120–5.
- Lipton R. B and Bigal M. E. The epidemiology of migraine. Am J Med. 2005;118:3S-10S.
- Liu A., Menon S., Colson N. J., Quinlan S., Cox H., Peterson M., Tiang T., Haupt L. M., Lea R. A and Griffiths L. R. Analysis of the MTHFR C677T variant with migraine phenotypes. BMC Res Notes. 2010;3:21.
- Oterino A., Valle N., Bravo Y., Munoz P., Sanchez-Velasco P., Ruiz- Alegrıa C., Castillo J., Leyva-Cobian F., Vadillo A and Pascual J. MTHFR T677 homozygosis influences the presence of aura in migraineurs. Cephalalgia. 2004;24:491–494.
- Ozbey U., Etem E., Ozel S and Berilgen M. S. Is There a Relation Between Insertion/Deletion Polymorphism of the Angiotensin Converting Enzyme Gene in Patients of Migraine with Aura and Migraine without Aura in Region of Eastern Turkey? Turkiye Klinikleri J Med Sci. 2010:30(2):502-6.
- Pandith A. A., Wani I. Y., Qasim I., Shah Z. A and Sheikh S. Evaluation of Risk Related to MTHFR 677C>T Gene Polymorphism in Migraine Patients in Kashmiri Population. Open Journal of Preventive Medicine. 2017;7:151-161.
- Paterna S., Di Pasquale P., Angelo D. A., Seidita G., Tuttolomondo A., Cardinale A., Tiziana M., Giuseppe F., Alfonso G., Marco T and Giuseppe L. Angiotensin-converting enzyme gene deletion polymorphism determines an increase in frequency of migraine attacks in patients suffering from migraine without aura. Eur Neurol. 2000;43(3):133–6.
- Piane M., Lulli P., Farinelli I., Simeoni S., De Filippis S., Patacchioli F. R and Martelletti P. Genetics of migraine and pharmacogenomics: some considerations.The journal of headache and pain. 2007;8(6):334–9.
- Pizza V., Infante F., Agresta A., Cassano D and Capasso A. Insertion/deletion polymorphism of the angiotensin I-converting enzyme gene in migraine patients. Phol. 2013;2:19-22.
- Raina J. K., Panjaliya R. K., Sharma M., Bhardwaj R., Bakaya A and Kumar P. Methylenetetrahydrofolate reductase C677T gene polymorphism and predisposition to essential hypertension. International Journal of Genetics. 2016;8(5):207-210.
- Raina J. K., Sharma M., Panjaliya R. K., Bhagat M., Sharma R., Bakaya A and Kumar P. Methylenetetrahydrofolate reductase C677T and methionine synthase A2756G gene polymorphisms and associated risk of cardiovascular diseases: A study from Jammu region. Indian heart journal. 2016;68(3):421-430.
- Rigat B., Hubert C., Alhenc-Gelas F., Cambien F., Corvol P., Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. Clin. Invest. 1990;86(4):1343-1346.
- Rubino E., Ferrero M., Rainero I., Binello E.,Vaula G and Pinessi L. Association of the C677T polymorphism in the MTHFR gene with migraine: a meta-analysis. Cephalalgia. 2009;29:818–825.
- Silberstein S. D. Migraine. Lancet. 2004;363:381-391.
- Sambrook J and Russell D. W. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA. 2001.
- Scher A. I., Terwindt G. M., Verschuren W. M., Kruit M. C., Blom H. J., Kowa H., Frants R. R., van den Maagdenberg A. M., van Buchem M., Ferrari M. D and Launer L. J. Migraine and MTHFR C677Tgenotype in a population based sample. Ann Neurol. 2006;59:372–375.
- Schurks M., Zee R. Y., Buring J. E and Kurth T. Interrelationships among the MTHFR 677C>T polymorphism, migraine, and cardiovascular disease. Neurology. 2008;71:505–513.
- Sezer S., Altınışık J., Bozkurt N., Akkanet S., Ateş O. Analysis Of Angiotensin Converting Enzyme (ACE) Gene Insertion/Deletion (I/D) Polymorphism In Migraine. Journal of Contemporary Medicine. 2013;3(1):7-11.
- Sharma P. Meta–analysis of the ACE gene in ischaemic stroke. J Neurol Neurosurg Psychiatry. 1998;64(2):227-230.
- Stuart S., Cox H. C., Lea R. A and Griffiths L. R. The role of the MTHFR gene in migraine. Headache. 2012;52:515–519.
- Tietjen G. E., Herial N. A., Utley C., White L., Yerga-Woolwine S and Joe B. Association of von Willebrand factor activity with ACE I/D and MTHFR C677T polymorphisms in migraine. Cephalalgia. 2009;29:960-968.
- Todt U., Freudenberg J., Goebel I., Netzer C., Heinze A., Heinze-Kuhn K., Gobel H and Kubisch C. MTHFR C677T polymorphism and migraine with aura. Ann Neurol. 2006;60:621–623.
- Tronvik E., Stovner L. J., Bovim G., White L. R., Gladwin A. J., Owen K and Schrader H. Angiotensin converting enzyme gene insertion/deletion polymorphism in migraine patients. BMC Neurol. 2008;8:4.
- Wald D. S.,Law M and Morris J. K. Homocysteine and cardiovascular disease: evidence on casuality from a meta-analysis. BMJ. 2002;325:1202.
- The World Health Report. Mental Health: New Understanding. New Hope. 2001. http://www.who.int/whr2001/.
- Yang M., Qiu C. C., Xu Q and Xiang H. D. Association of Angiotensin Converting Enzyme I/D Polymorphism with Type 2 Diabetes Mellitus1. Biomed. Environ. Sci. 2006;19:323-327.

This work is licensed under a Creative Commons Attribution 4.0 International License.





